1. Introduction to ESM3 Model Deployment
The Evolutionary Scale Modeling 3 (ESM3) model has emerged as a powerful tool for computational biology, capable of handling protein sequence prediction, structural analysis, and high-dimensional embeddings. While researchers and bioinformaticians widely use ESM3 for experimental purposes, deploying it in production environments opens up transformative possibilities for scalable, real-time applications in healthcare, drug discovery, and synthetic biology.
This chapter introduces the foundational concepts of deploying ESM3 models, highlights challenges, and sets the stage for building robust deployment workflows. It provides practical examples to clarify why production deployment is essential for maximizing ESM3’s utility.
1.1 Overview of ESM3 Models
What is ESM3?
ESM3 is a transformer-based model designed for protein sequence analysis. It excels in:
- Predicting secondary and tertiary structures.
- Generating embeddings to represent sequence relationships.
- Providing residue-level confidence scores for experimental validation.
Key Applications:
- Drug Discovery: Identifying binding sites or therapeutic targets.
- Synthetic Biology: Designing proteins with tailored properties.
- Environmental Science: Engineering enzymes for pollution degradation.
Why Deploy in Production?
- Scalability: Run analyses on thousands of sequences simultaneously.
- Real-Time Use: Support applications like diagnostics or live data monitoring.
- Reproducibility: Ensure consistent results across workflows and users.
Example Workflow:
- Input: A protein sequence in FASTA format.
- Processing: Use ESM3 to predict embeddings and structures.
- Output: Confidence scores and 3D structural data for downstream analysis.
1.2 Challenges in Deploying ESM3 Models
Although ESM3 is highly effective, deploying it in production environments presents unique challenges.
1. High Computational Demands
- Reason: ESM3 models are resource-intensive due to their large architecture and high-dimensional outputs.
- Impact: Running multiple sequences simultaneously can overwhelm local resources.
Solution:
- Leverage GPUs for inference to accelerate processing.
- Use mixed-precision inference to reduce memory usage without compromising accuracy.
2. Managing Large Data Outputs
- Reason: ESM3 generates high-dimensional embeddings and large structural files (e.g., PDB format).
- Impact: Managing and storing these outputs becomes cumbersome in large-scale projects.
Solution:
- Implement data pipelines for automated preprocessing and storage.
- Use cloud storage solutions like AWS S3 or Google Cloud Storage for scalability.
3. Ensuring Scalability
- Reason: A single-machine setup may not meet the needs of dynamic production workflows.
- Impact: Performance bottlenecks and limited scalability can hinder real-time applications.
Solution:
- Use container orchestration tools like Kubernetes to distribute workloads.
- Implement load balancers to handle varying levels of user requests.
4. Debugging and Monitoring
- Reason: Identifying issues in complex deployments can be time-consuming.
- Impact: Delays in resolving errors can disrupt workflows.
Solution:
- Set up logging and monitoring tools like Prometheus and Grafana for real-time diagnostics.
- Use structured error-handling mechanisms to prevent failures.
1.3 Goals and Scope of Deployment
This guide focuses on practical strategies to deploy ESM3 models in diverse environments. By the end, you’ll have the tools and knowledge to:
- Build scalable pipelines for handling ESM3 predictions.
- Optimize performance for batch and real-time inference.
- Address deployment challenges using modern DevOps practices.
1.4 Practical Example: Why Deploy in Production?
Consider a hypothetical scenario where a pharmaceutical company wants to analyze 1,000 protein sequences to identify potential drug targets. Let’s compare experimental usage with production deployment:
Experimental Setup:
- Workflow: Run ESM3 locally on a high-performance desktop.
- Challenges: Limited scalability, manual data handling, high risk of errors.
Production Deployment:
- Workflow: Deploy ESM3 in a Kubernetes cluster with GPU nodes.
- Advantages: Parallel processing of sequences, automated data pipelines, consistent outputs.
Steps in a Production Workflow:
- Input protein sequences into a centralized database.
- Trigger ESM3 predictions via an API for each sequence.
- Store predictions in a cloud-based storage system.
- Visualize results in a web dashboard for researchers.
Code Snippet for Batch Processing with ESM3:
pythonCopy codeimport torch
from esm import pretrained
# Load ESM3 model
model, alphabet = pretrained.esm3_t36_3B_UR50D()
batch_converter = alphabet.get_batch_converter()
# Batch processing
sequences = [
("Protein1", "MKTLLILAVVAAALA"),
("Protein2", "MKTLLIMVVVAAGLA"),
("Protein3", "MKTLLILAVIAAALA"),
]
batch_labels, batch_strs, batch_tokens = batch_converter(sequences)
# Inference
with torch.no_grad():
results = model(batch_tokens, repr_layers=[33])
embeddings = results["representations"][33]
print(f"Embedding for {sequences[0][0]}:", embeddings[0].shape)
1.5 The Road Ahead
This guide provides detailed, step-by-step instructions for deploying ESM3 models across different environments:
- Setting up hardware and software for local and cloud-based deployments.
- Containerizing ESM3 workflows for portability and reproducibility.
- Scaling deployments using Kubernetes and cloud infrastructure.
- Optimizing performance with techniques like mixed-precision inference.
- Ensuring security and compliance in production environments.
Each subsequent section will include comprehensive tutorials, real-world examples, and practical tips to ensure a smooth deployment process. By understanding the challenges and strategies for deployment, you’ll be ready to unlock the full potential of ESM3 in production workflows.
2. Setting Up the Deployment Environment
Deploying ESM3 models in production requires a well-prepared environment to handle the model’s computational demands and ensure efficient workflows. This chapter covers the hardware, software, and infrastructure setup necessary for deploying ESM3 models. It provides practical examples and step-by-step tutorials to create an optimized environment tailored to your specific deployment needs.
2.1 Hardware Requirements
ESM3 models are resource-intensive and require robust hardware for efficient operation, particularly when dealing with large datasets or real-time applications.
1. Local Deployment Hardware
A high-performance workstation is suitable for small-scale deployments or development.
Recommended Configuration:
- Processor: Intel i9 or AMD Ryzen 9.
- GPU: NVIDIA RTX 3080 or higher (CUDA compatibility required).
- Memory: Minimum 64 GB RAM.
- Storage: NVMe SSDs with at least 1 TB for fast read/write operations.
Example Scenario: A research lab uses a single workstation to analyze sequences in batches. The GPU accelerates predictions, while the SSD handles large output files.
2. Cloud Deployment Hardware
For larger workloads or distributed processing, cloud platforms are ideal. Providers like AWS, Google Cloud, and Azure offer GPU-accelerated instances.
Recommended Instances:
- AWS: g4dn.xlarge (1 NVIDIA T4 GPU, 16 GB GPU memory).
- Google Cloud: A2 High-GPU (1 NVIDIA A100 GPU, 40 GB GPU memory).
- Azure: NC6s_v3 (1 NVIDIA Tesla V100 GPU, 16 GB GPU memory).
Example Workflow: A pharmaceutical company deploys ESM3 on AWS to analyze 10,000 sequences simultaneously. Autoscaling ensures cost-effectiveness during low usage periods.
Launching an AWS GPU Instance:
bashCopy codeaws ec2 run-instances \
--instance-type g4dn.xlarge \
--image-id ami-0abcdef1234567890 \
--count 1 \
--key-name MyKeyPair \
--security-groups MySecurityGroup
3. Comparing Local vs. Cloud Hardware
| Feature | Local | Cloud |
|---|---|---|
| Initial Cost | High (hardware purchase) | Pay-as-you-go pricing |
| Scalability | Limited | Highly scalable |
| Maintenance | User-managed | Provider-managed |
| Latency | Low (local access) | May vary (network latency) |
2.2 Software Stack for Deployment
Setting up the correct software stack is crucial for deploying ESM3 models efficiently.
1. Operating System
- Recommended: Linux-based systems (e.g., Ubuntu 20.04) for better GPU compatibility and performance.
- Alternatives: Windows with WSL2 for Linux compatibility or macOS (without GPU support).
2. Required Libraries and Tools
Install essential software components for running ESM3:
- CUDA Toolkit: Enables GPU acceleration (minimum version: 11.3).
- PyTorch: Framework for running the ESM3 model.
- ESM Library: Pretrained ESM3 models.
- Additional Libraries: Matplotlib, NumPy, Pandas, Seaborn for data visualization and analysis.
3. Installing Dependencies
Step-by-Step Installation:
- Install CUDA Toolkit:bashCopy code
sudo apt update sudo apt install -y nvidia-cuda-toolkit nvidia-smi # Verify GPU availability - Set Up a Virtual Environment:bashCopy code
python3 -m venv esm3_env source esm3_env/bin/activate # Linux/Mac esm3_env\Scripts\activate # Windows - Install Python Libraries:bashCopy code
pip install torch torchvision torchaudio --extra-index-url https://download.pytorch.org/whl/cu113 pip install esm matplotlib seaborn pandas - Verify Installation:pythonCopy code
import torch print(f"CUDA available: {torch.cuda.is_available()}")
2.3 Infrastructure Planning
Selecting the right infrastructure depends on your deployment goals. Consider the following setups:
1. Single-Machine Setup
Ideal for development or small-scale use.
Workflow:
- Preprocess sequences locally.
- Run ESM3 on a GPU-enabled workstation.
- Store outputs on local drives.
Benefits:
- Low latency for local access.
- Minimal setup time.
2. Distributed Systems
For larger workloads, use distributed systems to handle multiple tasks concurrently.
Key Tools:
- HPC Clusters: Use Slurm for managing batch jobs in high-performance computing environments.
- Cloud Platforms: AWS Batch, GCP AI Platform, or Azure Machine Learning.
Example: Running Batch Jobs with Slurm:
bashCopy code#!/bin/bash
#SBATCH --job-name=esm3_job
#SBATCH --ntasks=1
#SBATCH --gpus=1
#SBATCH --time=04:00:00
#SBATCH --output=esm3_output.log
module load cuda/11.3
python run_esm3.py --input sequences.fasta --output results.json
Submit the job:
bashCopy codesbatch esm3_job.sh
3. Hybrid Infrastructure
Combine local and cloud resources for flexibility:
- Use local machines for testing and development.
- Deploy production workflows on the cloud for scalability.
2.4 Practical Example: Setting Up an Environment
Scenario:
A bioinformatics lab wants to process protein sequences with ESM3 on a local GPU-enabled workstation.
Steps:
- Install CUDA and PyTorch:bashCopy code
sudo apt install -y nvidia-cuda-toolkit pip install torch torchvision torchaudio --extra-index-url https://download.pytorch.org/whl/cu113 - Download and Verify ESM3 Model:pythonCopy code
from esm import pretrained model, alphabet = pretrained.esm3_t36_3B_UR50D() print("Model loaded successfully") - Run Test Inference:pythonCopy code
sequence = "MKTLLILAVVAAALA" batch_converter = alphabet.get_batch_converter() batch_labels, batch_strs, batch_tokens = batch_converter([("Test", sequence)]) with torch.no_grad(): result = model(batch_tokens, repr_layers=[33]) print("Embedding shape:", result["representations"][33].shape) - Benchmark GPU Utilization:bashCopy code
nvidia-smi
This chapter has detailed the hardware, software, and infrastructure required to deploy ESM3 models. By setting up an efficient environment tailored to your needs, you can ensure a smooth and scalable deployment process. The next chapter will focus on containerizing ESM3 models for portable and reproducible workflows.
3. Containerizing ESM3 Models
Containerization is a vital step in deploying ESM3 models, ensuring portability, reproducibility, and ease of deployment across diverse environments. By encapsulating the model, dependencies, and configurations into a container, you can run ESM3 workflows consistently, whether on local machines, cloud platforms, or high-performance clusters.
This chapter provides a comprehensive guide to containerizing ESM3 models using Docker. It includes practical examples, debugging tips, and strategies for deploying containers across environments.
3.1 Introduction to Containers
What Are Containers?
Containers are lightweight, standalone software packages that include all necessary dependencies, libraries, and configurations to run an application.
Why Use Containers for ESM3?
- Portability: Run ESM3 workflows seamlessly across different systems.
- Reproducibility: Ensure consistent results by packaging the exact runtime environment.
- Ease of Deployment: Simplify deployment on cloud platforms or Kubernetes clusters.
Containerization Tools:
- Docker: Most widely used containerization tool.
- Singularity: Ideal for high-performance computing (HPC) environments.
3.2 Writing a Dockerfile for ESM3
The Dockerfile is the blueprint for creating a container. Below, we create a Dockerfile optimized for running ESM3 workflows.
Basic Structure of a Dockerfile:
- Base Image: Start with a prebuilt image like Python or CUDA.
- Install Dependencies: Add Python libraries, CUDA, and ESM3.
- Add Model Code: Copy ESM3 scripts or models into the container.
- Set Entry Point: Define how the container should run.
Step-by-Step Dockerfile for ESM3:
dockerfileCopy code# Base Image: Use a CUDA-enabled image for GPU support
FROM nvidia/cuda:11.3.1-base-ubuntu20.04
# Set environment variables for Python
ENV PYTHONUNBUFFERED=1 \
DEBIAN_FRONTEND=noninteractive
# Install system dependencies
RUN apt-get update && apt-get install -y \
python3 python3-pip git curl wget
# Set up Python environment
RUN python3 -m pip install --upgrade pip \
&& pip install torch torchvision torchaudio --extra-index-url https://download.pytorch.org/whl/cu113 \
&& pip install esm matplotlib seaborn pandas
# Add ESM3 script to the container
WORKDIR /app
COPY run_esm3.py /app/
# Set default command
CMD ["python3", "run_esm3.py"]
Explaining the Steps:
- Base Image: The NVIDIA CUDA image ensures GPU support for ESM3.
- System Dependencies: Install Python, pip, and utilities like Git and Curl.
- Python Libraries: Install PyTorch (with CUDA) and the ESM library.
- Application Code: Copy ESM3-related scripts into the container.
3.3 Building and Running the Container
Building the Docker Image:
Use the docker build command to create an image from the Dockerfile.
bashCopy codedocker build -t esm3-container .
Command Breakdown:
-t esm3-container: Tags the image with the nameesm3-container..: Specifies the current directory as the build context.
Running the Docker Container:
Launch the container and execute the default command defined in the Dockerfile.
bashCopy codedocker run --gpus all esm3-container
Command Breakdown:
--gpus all: Enables GPU access for the container.esm3-container: Specifies the container image to run.
Testing GPU Support:
Verify that the container has access to the GPU by running nvidia-smi.
bashCopy codedocker run --gpus all nvidia/cuda:11.3.1-base-ubuntu20.04 nvidia-smi
Output:
luaCopy code+-----------------------------------------------------------------------------+
| NVIDIA-SMI 470.57.02 Driver Version: 470.57.02 CUDA Version: 11.3 |
|-------------------------------+----------------------+----------------------+
3.4 Debugging Common Issues
1. Missing GPU Support:
- Symptom: The container runs but cannot access the GPU.
- Solution: Ensure the
nvidia-container-toolkitis installed and properly configured.bashCopy codesudo apt-get install -y nvidia-container-toolkit sudo systemctl restart docker
2. Dependency Conflicts:
- Symptom: Errors during package installation (e.g., PyTorch version mismatch).
- Solution: Match PyTorch and CUDA versions by referencing the PyTorch compatibility matrix.
3. Large Image Size:
- Symptom: Docker image exceeds several GBs.
- Solution:
- Use
slimbase images (e.g.,python:3.8-slim). - Avoid unnecessary dependencies.
- Use
3.5 Advanced Dockerfile Features
1. Multi-Stage Builds:
Reduce image size by separating build and runtime dependencies.
dockerfileCopy code# Stage 1: Build
FROM python:3.8-slim as builder
RUN pip install torch esm
# Stage 2: Runtime
FROM nvidia/cuda:11.3.1-base-ubuntu20.04
COPY --from=builder /usr/local/lib/python3.8/dist-packages /usr/local/lib/python3.8/dist-packages
2. Custom Entrypoints:
Allow dynamic execution of different scripts.
dockerfileCopy codeENTRYPOINT ["python3"]
CMD ["run_esm3.py"]
Run the container with a different script:
bashCopy codedocker run esm3-container another_script.py
3.6 Deploying Docker Containers
1. Hosting on Docker Hub:
Share your container image by pushing it to Docker Hub.
bashCopy codedocker tag esm3-container mydockerhub/esm3-container
docker push mydockerhub/esm3-container
2. Running on Cloud Platforms:
Deploy the Docker container on cloud services like AWS, Google Cloud, or Azure.
AWS Elastic Container Service (ECS):
- Push the Docker image to Amazon Elastic Container Registry (ECR).bashCopy code
aws ecr create-repository --repository-name esm3-container docker tag esm3-container <ecr-repo-uri> docker push <ecr-repo-uri> - Create an ECS task definition using the container.
3.7 Using Singularity for HPC
For environments like academic clusters, Singularity is preferred over Docker due to its compatibility with shared systems.
Convert Docker to Singularity:
Use the docker2singularity tool.
bashCopy codesingularity build esm3.sif docker://esm3-container
Run the Singularity Container:
bashCopy codesingularity exec esm3.sif python3 run_esm3.py
3.8 Practical Example: End-to-End Workflow
Scenario:
A research lab wants to containerize and deploy ESM3 on a Kubernetes cluster for batch protein analysis.
Steps:
- Write the Dockerfile: Follow the steps in 3.2 to create and test the
esm3-container. - Push the Container to Docker Hub:bashCopy code
docker tag esm3-container mydockerhub/esm3-container docker push mydockerhub/esm3-container - Deploy on Kubernetes: Create a deployment YAML file:yamlCopy code
apiVersion: apps/v1 kind: Deployment metadata: name: esm3-deployment spec: replicas: 2 selector: matchLabels: app: esm3 template: metadata: labels: app: esm3 spec: containers: - name: esm3 image: mydockerhub/esm3-container resources: limits: nvidia.com/gpu: 1 - Apply the Deployment:bashCopy code
kubectl apply -f esm3-deployment.yaml
This chapter provided a detailed guide to containerizing ESM3 models, covering Dockerfile creation, debugging, and deployment strategies. By using containers, you can ensure portability, scalability, and reproducibility in your ESM3 workflows. The next chapter will focus on orchestrating these containers with Kubernetes for large-scale deployments.
4. Deploying ESM3 with Kubernetes
Kubernetes is the go-to platform for orchestrating containers in production environments. By deploying ESM3 with Kubernetes, you gain scalability, reliability, and efficient resource management for handling large-scale workloads. This chapter explains how to deploy ESM3 using Kubernetes, from setting up a cluster to creating deployments and managing workflows.
4.1 Introduction to Kubernetes
Kubernetes is an open-source platform that automates the deployment, scaling, and management of containerized applications.
Key Kubernetes Components:
- Pods: The smallest deployable unit that encapsulates containers.
- Nodes: Machines (virtual or physical) that run the containers.
- Deployments: Controllers that manage the desired state of pods.
- Services: Provide networking to expose applications to internal or external clients.
Why Use Kubernetes for ESM3?
- Scalability: Run multiple instances of ESM3 to handle large workloads.
- Resilience: Automatically restart failed pods.
- Load Balancing: Distribute requests across containers efficiently.
- Flexibility: Integrate with GPUs and scale dynamically based on demand.
4.2 Setting Up Kubernetes for ESM3
1. Installing Kubernetes
Kubernetes can be installed on various environments, such as local machines, cloud platforms, or on-premises servers.
Local Setup:
- Use Minikube: Ideal for development and testing.bashCopy code
curl -LO https://storage.googleapis.com/minikube/releases/latest/minikube-linux-amd64 sudo install minikube-linux-amd64 /usr/local/bin/minikube minikube start --driver=docker - Verify Installation:bashCopy code
kubectl get nodes
Cloud-Based Setup:
- Use a Managed Kubernetes Service:
- AWS: Elastic Kubernetes Service (EKS)
- GCP: Google Kubernetes Engine (GKE)
- Azure: Azure Kubernetes Service (AKS)
Example: Creating an EKS Cluster:
bashCopy codeeksctl create cluster --name esm3-cluster --nodes 3 --region us-west-2
2. Configuring GPU Support
GPU nodes are essential for running ESM3 efficiently.
- Install the NVIDIA device plugin:bashCopy code
kubectl apply -f https://raw.githubusercontent.com/NVIDIA/k8s-device-plugin/main/nvidia-device-plugin.yml - Verify GPU availability:bashCopy code
kubectl get nodes "-o=custom-columns=NAME:.metadata.name,GPUS:.status.allocatable.nvidia\.com/gpu"
3. Setting Up Kubernetes CLI
Ensure that kubectl is installed and configured for managing the cluster:
bashCopy codesudo apt-get install -y kubectl
kubectl config view # Check configuration
4.3 Deploying ESM3 with Kubernetes
Step 1: Writing the Deployment YAML
The YAML file defines the configuration for deploying ESM3 containers on Kubernetes.
Basic Deployment YAML for ESM3:
yamlCopy codeapiVersion: apps/v1
kind: Deployment
metadata:
name: esm3-deployment
spec:
replicas: 2 # Number of container instances
selector:
matchLabels:
app: esm3
template:
metadata:
labels:
app: esm3
spec:
containers:
- name: esm3
image: mydockerhub/esm3-container:latest
resources:
limits:
nvidia.com/gpu: 1 # Allocate one GPU per container
ports:
- containerPort: 5000 # Port for API access
Explanation of Key Sections:
- replicas: Defines the number of instances of the ESM3 container.
- image: Specifies the Docker image of the container.
- resources: Allocates GPU resources to the container.
- containerPort: Exposes the application inside the container.
Step 2: Creating the Deployment
Apply the deployment configuration:
bashCopy codekubectl apply -f esm3-deployment.yaml
Verify the deployment:
bashCopy codekubectl get pods
Step 3: Exposing the Service
Expose the deployment to allow external access.
Service YAML:
yamlCopy codeapiVersion: v1
kind: Service
metadata:
name: esm3-service
spec:
type: LoadBalancer
selector:
app: esm3
ports:
- protocol: TCP
port: 80
targetPort: 5000
Apply the service:
bashCopy codekubectl apply -f esm3-service.yaml
Get the external IP of the service:
bashCopy codekubectl get service esm3-service
4.4 Scaling ESM3 with Kubernetes
Kubernetes makes it easy to scale applications based on workload.
Manual Scaling: Increase or decrease the number of replicas:
bashCopy codekubectl scale deployment esm3-deployment --replicas=5
Autoscaling: Enable horizontal pod autoscaling:
bashCopy codekubectl autoscale deployment esm3-deployment --cpu-percent=80 --min=2 --max=10
View autoscaler status:
bashCopy codekubectl get hpa
4.5 Monitoring and Debugging Kubernetes Deployments
Monitoring Tools:
- Kubernetes Dashboard: A web-based interface for cluster management.bashCopy code
minikube dashboard - Prometheus and Grafana: For advanced metrics and visualization.
Debugging Tools:
- Inspect Logs:bashCopy code
kubectl logs <pod-name> - Check Pod Details:bashCopy code
kubectl describe pod <pod-name> - Debugging Failures:bashCopy code
kubectl get events
4.6 Practical Example: End-to-End Kubernetes Workflow
Scenario: A bioinformatics company wants to deploy ESM3 on Kubernetes to process batch protein sequences.
Steps:
- Create the Deployment: Write and apply the
esm3-deployment.yamlfile. - Scale the Deployment: Autoscale the deployment based on CPU usage.
- Expose the Service: Use the
esm3-service.yamlfile to allow external access. - Monitor the Deployment: Use
kubectl logsand Prometheus to track resource usage and request patterns. - Test the API: Send a test sequence to the exposed API:bashCopy code
curl -X POST -H "Content-Type: application/json" \ -d '{"sequence": "MKTLLILAVVAAALA"}' http://<external-ip>/predict
Deploying ESM3 with Kubernetes provides a scalable, resilient, and efficient solution for production workloads. This chapter outlined the setup process, from cluster configuration to deploying, scaling, and monitoring ESM3. The next chapter will focus on building data pipelines to automate preprocessing, predictions, and result handling.
5. Building Data Pipelines for ESM3
Effective data pipelines are essential for automating workflows in production environments. ESM3 requires streamlined preprocessing, inference, and postprocessing to handle large-scale datasets efficiently. This chapter explores how to design, implement, and optimize data pipelines tailored to ESM3, with detailed tutorials and examples.
5.1 Data Preprocessing for Production
Preprocessing ensures input data is correctly formatted for ESM3. Common tasks include cleaning protein sequences, validating input files, and organizing data into batches.
1. Validating Input Data
Ensure input sequences conform to the FASTA format and contain valid amino acid characters.
Example: Validation Script
pythonCopy codeimport re
def validate_fasta(sequence):
valid_chars = re.compile("^[ACDEFGHIKLMNPQRSTVWY]+![Rendered by QuickLaTeX.com ") if not valid_chars.match(sequence): raise ValueError("Invalid characters in sequence.") return True # Example usage sequence = "MKTLLILAVVAAALA" if validate_fasta(sequence): print("Sequence is valid.") </code></pre> <!-- /wp:preformatted --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:paragraph --> <strong>2. Cleaning Input Data</strong> <!-- /wp:paragraph --> <!-- wp:paragraph --> Remove invalid sequences, duplicates, or entries with missing data. <!-- /wp:paragraph --> <!-- wp:paragraph --> <strong>Example: Cleaning FASTA Files</strong> <!-- /wp:paragraph --> <!-- wp:preformatted --> <pre class="wp-block-preformatted">pythonCopy code<code>def clean_fasta(input_file, output_file): valid_sequences = [] with open(input_file, "r") as f: for line in f: if line.startswith(">"): header = line.strip() else: sequence = line.strip() if validate_fasta(sequence): valid_sequences.append((header, sequence)) with open(output_file, "w") as f_out: for header, seq in valid_sequences: f_out.write(f"{header}\n{seq}\n") clean_fasta("raw_sequences.fasta", "cleaned_sequences.fasta") </code></pre> <!-- /wp:preformatted --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:paragraph --> <strong>3. Batch Preparation</strong> <!-- /wp:paragraph --> <!-- wp:paragraph --> Batching sequences improves inference efficiency, especially in GPU environments. <!-- /wp:paragraph --> <!-- wp:paragraph --> <strong>Batching Example:</strong> <!-- /wp:paragraph --> <!-- wp:preformatted --> <pre class="wp-block-preformatted">pythonCopy code<code>def create_batches(sequences, batch_size): for i in range(0, len(sequences), batch_size): yield sequences[i:i+batch_size] # Example usage sequences = ["MKTLLILAVVAAALA", "MKTLLIMVVVAAGLA", "MKTLLILAVIAAALA"] for batch in create_batches(sequences, batch_size=2): print(batch) </code></pre> <!-- /wp:preformatted --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>5.2 Workflow Automation</strong></h4> <!-- /wp:heading --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:paragraph --> Automation tools like Apache Airflow and Prefect enable scheduling and managing data pipelines. <!-- /wp:paragraph --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:paragraph --> <strong>1. Introduction to Airflow</strong> <!-- /wp:paragraph --> <!-- wp:paragraph --> Apache Airflow is a workflow orchestration tool for managing tasks like data preprocessing and model inference. <!-- /wp:paragraph --> <!-- wp:paragraph --> <strong>Installing Airflow:</strong> <!-- /wp:paragraph --> <!-- wp:preformatted --> <pre class="wp-block-preformatted">bashCopy code<code>pip install apache-airflow </code></pre> <!-- /wp:preformatted --> <!-- wp:paragraph --> <strong>Basic Airflow DAG:</strong> <!-- /wp:paragraph --> <!-- wp:preformatted --> <pre class="wp-block-preformatted">pythonCopy code<code>from airflow import DAG from airflow.operators.python_operator import PythonOperator from datetime import datetime def preprocess_data(): print("Preprocessing data...") def run_inference(): print("Running ESM3 inference...") default_args = { "owner": "airflow", "start_date": datetime(2023, 1, 1), "retries": 1, } with DAG("esm3_pipeline", default_args=default_args, schedule_interval="@daily") as dag: preprocess_task = PythonOperator(task_id="preprocess_data", python_callable=preprocess_data) inference_task = PythonOperator(task_id="run_inference", python_callable=run_inference) preprocess_task >> inference_task </code></pre> <!-- /wp:preformatted --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:paragraph --> <strong>2. Automating with Prefect</strong> <!-- /wp:paragraph --> <!-- wp:paragraph --> Prefect offers a more Pythonic and flexible approach to building workflows. <!-- /wp:paragraph --> <!-- wp:paragraph --> <strong>Basic Prefect Flow:</strong> <!-- /wp:paragraph --> <!-- wp:preformatted --> <pre class="wp-block-preformatted">pythonCopy code<code>from prefect import Flow, task @task def preprocess_data(): print("Preprocessing data...") @task def run_inference(): print("Running ESM3 inference...") with Flow("esm3_pipeline") as flow: data = preprocess_data() run_inference(upstream_tasks=[data]) flow.run() </code></pre> <!-- /wp:preformatted --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>5.3 Data Management and Storage</strong></h4> <!-- /wp:heading --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:paragraph --> Managing ESM3 outputs, such as embeddings and predictions, requires robust storage solutions. <!-- /wp:paragraph --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:paragraph --> <strong>1. File-Based Storage</strong> <!-- /wp:paragraph --> <!-- wp:paragraph --> Store outputs as JSON or CSV files for easy retrieval. <!-- /wp:paragraph --> <!-- wp:paragraph --> <strong>Example: Saving Predictions to JSON</strong> <!-- /wp:paragraph --> <!-- wp:preformatted --> <pre class="wp-block-preformatted">pythonCopy code<code>import json predictions = {"sequence": "MKTLLILAVVAAALA", "confidence_scores": [0.95, 0.89, 0.88]} with open("predictions.json", "w") as f: json.dump(predictions, f) </code></pre> <!-- /wp:preformatted --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:paragraph --> <strong>2. Cloud Storage</strong> <!-- /wp:paragraph --> <!-- wp:paragraph --> For scalable storage, use cloud services like AWS S3 or Google Cloud Storage. <!-- /wp:paragraph --> <!-- wp:paragraph --> <strong>Example: Uploading to AWS S3</strong> <!-- /wp:paragraph --> <!-- wp:preformatted --> <pre class="wp-block-preformatted">bashCopy code<code>aws s3 cp predictions.json s3://my-esm3-bucket/predictions/ </code></pre> <!-- /wp:preformatted --> <!-- wp:paragraph --> <strong>Python Script for Uploading:</strong> <!-- /wp:paragraph --> <!-- wp:preformatted --> <pre class="wp-block-preformatted">pythonCopy code<code>import boto3 s3 = boto3.client("s3") s3.upload_file("predictions.json", "my-esm3-bucket", "predictions/predictions.json") </code></pre> <!-- /wp:preformatted --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:paragraph --> <strong>3. Database Storage</strong> <!-- /wp:paragraph --> <!-- wp:paragraph --> Store predictions in a database for querying and integration with applications. <!-- /wp:paragraph --> <!-- wp:paragraph --> <strong>Using SQLite to Store Predictions:</strong> <!-- /wp:paragraph --> <!-- wp:preformatted --> <pre class="wp-block-preformatted">pythonCopy code<code>import sqlite3 # Create a database and table conn = sqlite3.connect("esm3_predictions.db") cursor = conn.cursor() cursor.execute(""" CREATE TABLE IF NOT EXISTS predictions ( id INTEGER PRIMARY KEY, sequence TEXT, confidence_scores TEXT ) """) conn.commit() # Insert data sequence = "MKTLLILAVVAAALA" confidence_scores = "[0.95, 0.89, 0.88]" cursor.execute("INSERT INTO predictions (sequence, confidence_scores) VALUES (?, ?)", (sequence, confidence_scores)) conn.commit() # Query data cursor.execute("SELECT * FROM predictions") print(cursor.fetchall()) </code></pre> <!-- /wp:preformatted --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>5.4 Practical Example: End-to-End Pipeline</strong></h4> <!-- /wp:heading --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:paragraph --> <strong>Scenario:</strong> Automate the pipeline for processing protein sequences, running ESM3 predictions, and saving outputs. <!-- /wp:paragraph --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:paragraph --> <strong>Pipeline Steps:</strong> <!-- /wp:paragraph --> <!-- wp:list {"ordered":true} --> <ol class="wp-block-list"><!-- wp:list-item --> <li>Preprocess raw sequences.</li> <!-- /wp:list-item --> <!-- wp:list-item --> <li>Batch sequences for inference.</li> <!-- /wp:list-item --> <!-- wp:list-item --> <li>Run predictions using ESM3.</li> <!-- /wp:list-item --> <!-- wp:list-item --> <li>Save predictions to a database.</li> <!-- /wp:list-item --></ol> <!-- /wp:list --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:paragraph --> <strong>Complete Python Script:</strong> <!-- /wp:paragraph --> <!-- wp:preformatted --> <pre class="wp-block-preformatted">pythonCopy code<code>import sqlite3 from esm import pretrained # Step 1: Preprocess Sequences def preprocess_sequences(input_file): sequences = [] with open(input_file, "r") as f: for line in f: if not line.startswith(">"): sequences.append(line.strip()) return sequences # Step 2: Batch Sequences def create_batches(sequences, batch_size): for i in range(0, len(sequences), batch_size): yield sequences[i:i+batch_size] # Step 3: Run ESM3 Predictions def run_esm3(sequences): model, alphabet = pretrained.esm3_t36_3B_UR50D() batch_converter = alphabet.get_batch_converter() batch_labels, batch_strs, batch_tokens = batch_converter([(str(i), seq) for i, seq in enumerate(sequences)]) with torch.no_grad(): results = model(batch_tokens, repr_layers=[33]) embeddings = results["representations"][33] return embeddings # Step 4: Save Outputs def save_to_db(sequence, embeddings): conn = sqlite3.connect("esm3_predictions.db") cursor = conn.cursor() cursor.execute(""" CREATE TABLE IF NOT EXISTS predictions ( id INTEGER PRIMARY KEY, sequence TEXT, embeddings TEXT ) """) conn.commit() cursor.execute("INSERT INTO predictions (sequence, embeddings) VALUES (?, ?)", (sequence, str(embeddings.tolist()))) conn.commit() conn.close() # Main Pipeline def main(): sequences = preprocess_sequences("sequences.fasta") for batch in create_batches(sequences, batch_size=5): embeddings = run_esm3(batch) for seq, emb in zip(batch, embeddings): save_to_db(seq, emb) main() </code></pre> <!-- /wp:preformatted --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:paragraph --> This chapter detailed how to build and automate data pipelines for ESM3 workflows. From preprocessing sequences to storing outputs in databases or cloud storage, these steps ensure efficient and scalable workflows. The next chapter will focus on serving ESM3 models through APIs for real-time and batch predictions. <!-- /wp:paragraph --> <!-- wp:heading {"level":3} --> <h3 class="wp-block-heading"><strong>6. Serving ESM3 Models Through APIs</strong></h3> <!-- /wp:heading --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:paragraph --> Serving ESM3 models via APIs enables real-time and batch predictions for end-users and applications. This chapter focuses on setting up a RESTful API using Python's <strong>FastAPI</strong> framework. We'll cover step-by-step instructions for building, deploying, and optimizing an API for ESM3, complete with practical examples. <!-- /wp:paragraph --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>6.1 Introduction to FastAPI</strong></h4> <!-- /wp:heading --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:paragraph --> FastAPI is a high-performance web framework for building APIs, leveraging Python's type hints for validation and auto-generating documentation. <!-- /wp:paragraph --> <!-- wp:paragraph --> <strong>Why Use FastAPI for ESM3?</strong> <!-- /wp:paragraph --> <!-- wp:list {"ordered":true} --> <ol class="wp-block-list"><!-- wp:list-item --> <li><strong>Ease of Use:</strong> Simple syntax for defining endpoints.</li> <!-- /wp:list-item --> <!-- wp:list-item --> <li><strong>Performance:</strong> Built on ASGI, capable of handling high-concurrency requests.</li> <!-- /wp:list-item --> <!-- wp:list-item --> <li><strong>Documentation:</strong> Automatically generates interactive Swagger UI.</li> <!-- /wp:list-item --></ol> <!-- /wp:list --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:paragraph --> <strong>Installing FastAPI:</strong> <!-- /wp:paragraph --> <!-- wp:preformatted --> <pre class="wp-block-preformatted">bashCopy code<code>pip install fastapi uvicorn </code></pre> <!-- /wp:preformatted --> <!-- wp:paragraph --> <strong>Starting a FastAPI Server:</strong> Create a simple API to verify the setup: <!-- /wp:paragraph --> <!-- wp:preformatted --> <pre class="wp-block-preformatted">pythonCopy code<code>from fastapi import FastAPI app = FastAPI() @app.get("/") def read_root(): return {"message": "Welcome to the ESM3 API"} </code></pre> <!-- /wp:preformatted --> <!-- wp:paragraph --> Run the server: <!-- /wp:paragraph --> <!-- wp:preformatted --> <pre class="wp-block-preformatted">bashCopy code<code>uvicorn main:app --host 0.0.0.0 --port 8000 </code></pre> <!-- /wp:preformatted --> <!-- wp:paragraph --> Visit <a>http://localhost:8000/docs</a> to access the interactive documentation. <!-- /wp:paragraph --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>6.2 Building an API for ESM3</strong></h4> <!-- /wp:heading --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:paragraph --> <strong>Step 1: Define the API Structure</strong> <!-- /wp:paragraph --> <!-- wp:paragraph --> Plan the API endpoints: <!-- /wp:paragraph --> <!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li><strong><code>/predict</code> (POST):</strong> Accepts sequences for ESM3 prediction.</li> <!-- /wp:list-item --> <!-- wp:list-item --> <li><strong><code>/batch_predict</code> (POST):</strong> Processes multiple sequences in a batch.</li> <!-- /wp:list-item --> <!-- wp:list-item --> <li><strong><code>/healthcheck</code> (GET):</strong> Monitors API status.</li> <!-- /wp:list-item --></ul> <!-- /wp:list --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:paragraph --> <strong>Step 2: Setting Up the Prediction Endpoint</strong> <!-- /wp:paragraph --> <!-- wp:paragraph --> Create a <code>predict</code> endpoint for single-sequence predictions. <!-- /wp:paragraph --> <!-- wp:paragraph --> <strong>Example Code:</strong> <!-- /wp:paragraph --> <!-- wp:preformatted --> <pre class="wp-block-preformatted">pythonCopy code<code>from fastapi import FastAPI, HTTPException from pydantic import BaseModel import torch from esm import pretrained # Initialize FastAPI app app = FastAPI() # Load ESM3 model at startup model, alphabet = pretrained.esm3_t36_3B_UR50D() batch_converter = alphabet.get_batch_converter() # Define input schema class SequenceInput(BaseModel): sequence: str @app.post("/predict") def predict(input_data: SequenceInput): sequence = input_data.sequence.upper() # Validate sequence valid_chars = set("ACDEFGHIKLMNPQRSTVWY") if not all(c in valid_chars for c in sequence): raise HTTPException(status_code=400, detail="Invalid sequence characters") # Run ESM3 prediction batch_labels, batch_strs, batch_tokens = batch_converter([("input", sequence)]) with torch.no_grad(): result = model(batch_tokens, repr_layers=[33]) embedding = result["representations"][33][0].tolist() return {"sequence": sequence, "embedding": embedding} </code></pre> <!-- /wp:preformatted --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:paragraph --> <strong>Step 3: Adding Batch Processing</strong> <!-- /wp:paragraph --> <!-- wp:paragraph --> Handle multiple sequences in a single request with the <code>/batch_predict</code> endpoint. <!-- /wp:paragraph --> <!-- wp:paragraph --> <strong>Batch Prediction Endpoint:</strong> <!-- /wp:paragraph --> <!-- wp:preformatted --> <pre class="wp-block-preformatted">pythonCopy code<code>from typing import List class BatchInput(BaseModel): sequences: List[str] @app.post("/batch_predict") def batch_predict(input_data: BatchInput): sequences = [seq.upper() for seq in input_data.sequences] # Validate sequences valid_chars = set("ACDEFGHIKLMNPQRSTVWY") for seq in sequences: if not all(c in valid_chars for c in seq): raise HTTPException(status_code=400, detail=f"Invalid characters in sequence: {seq}") # Run predictions in batches results = [] for seq in sequences: batch_labels, batch_strs, batch_tokens = batch_converter([("input", seq)]) with torch.no_grad(): result = model(batch_tokens, repr_layers=[33]) embedding = result["representations"][33][0].tolist() results.append({"sequence": seq, "embedding": embedding}) return {"results": results} </code></pre> <!-- /wp:preformatted --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:paragraph --> <strong>Step 4: Adding Health Check Endpoint</strong> <!-- /wp:paragraph --> <!-- wp:paragraph --> Ensure the API is running properly with a <code>/healthcheck</code> endpoint. <!-- /wp:paragraph --> <!-- wp:paragraph --> <strong>Health Check Endpoint:</strong> <!-- /wp:paragraph --> <!-- wp:preformatted --> <pre class="wp-block-preformatted">pythonCopy code<code>@app.get("/healthcheck") def healthcheck(): return {"status": "healthy", "model_loaded": True} </code></pre> <!-- /wp:preformatted --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>6.3 Running and Testing the API</strong></h4> <!-- /wp:heading --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:paragraph --> <strong>Starting the Server:</strong> <!-- /wp:paragraph --> <!-- wp:preformatted --> <pre class="wp-block-preformatted">bashCopy code<code>uvicorn main:app --host 0.0.0.0 --port 8000 </code></pre> <!-- /wp:preformatted --> <!-- wp:paragraph --> <strong>Testing with cURL:</strong> <!-- /wp:paragraph --> <!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Single Prediction:bashCopy code<code>curl -X POST "http://localhost:8000/predict" -H "Content-Type: application/json" \ -d '{"sequence": "MKTLLILAVVAAALA"}'</code></li> <!-- /wp:list-item --> <!-- wp:list-item --> <li>Batch Prediction:bashCopy code<code>curl -X POST "http://localhost:8000/batch_predict" -H "Content-Type: application/json" \ -d '{"sequences": ["MKTLLILAVVAAALA", "ACDEFGHIKLMNPQRSTVWY"]}'</code></li> <!-- /wp:list-item --></ul> <!-- /wp:list --> <!-- wp:paragraph --> <strong>Output Example:</strong> <!-- /wp:paragraph --> <!-- wp:preformatted --> <pre class="wp-block-preformatted">jsonCopy code<code>{ "sequence": "MKTLLILAVVAAALA", "embedding": [0.12, 0.45, 0.33, ...] } </code></pre> <!-- /wp:preformatted --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>6.4 Deploying the API</strong></h4> <!-- /wp:heading --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:paragraph --> <strong>1. Containerizing the API</strong> <!-- /wp:paragraph --> <!-- wp:paragraph --> Write a <code>Dockerfile</code> for the FastAPI server: <!-- /wp:paragraph --> <!-- wp:preformatted --> <pre class="wp-block-preformatted">dockerfileCopy code<code>FROM python:3.9-slim # Install dependencies RUN pip install fastapi uvicorn torch esm # Copy application code WORKDIR /app COPY main.py /app/ # Run the server CMD ["uvicorn", "main:app", "--host", "0.0.0.0", "--port", "8000"] </code></pre> <!-- /wp:preformatted --> <!-- wp:paragraph --> Build and run the container: <!-- /wp:paragraph --> <!-- wp:preformatted --> <pre class="wp-block-preformatted">bashCopy code<code>docker build -t esm3-api . docker run -p 8000:8000 esm3-api </code></pre> <!-- /wp:preformatted --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:paragraph --> <strong>2. Deploying on Kubernetes</strong> <!-- /wp:paragraph --> <!-- wp:paragraph --> Write a deployment YAML: <!-- /wp:paragraph --> <!-- wp:preformatted --> <pre class="wp-block-preformatted">yamlCopy code<code>apiVersion: apps/v1 kind: Deployment metadata: name: esm3-api-deployment spec: replicas: 2 selector: matchLabels: app: esm3-api template: metadata: labels: app: esm3-api spec: containers: - name: esm3-api image: esm3-api:latest ports: - containerPort: 8000 </code></pre> <!-- /wp:preformatted --> <!-- wp:paragraph --> Expose the service: <!-- /wp:paragraph --> <!-- wp:preformatted --> <pre class="wp-block-preformatted">yamlCopy code<code>apiVersion: v1 kind: Service metadata: name: esm3-api-service spec: type: LoadBalancer selector: app: esm3-api ports: - protocol: TCP port: 80 targetPort: 8000 </code></pre> <!-- /wp:preformatted --> <!-- wp:paragraph --> Deploy and test: <!-- /wp:paragraph --> <!-- wp:preformatted --> <pre class="wp-block-preformatted">bashCopy code<code>kubectl apply -f esm3-api-deployment.yaml kubectl apply -f esm3-api-service.yaml </code></pre> <!-- /wp:preformatted --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>6.5 Optimizing the API</strong></h4> <!-- /wp:heading --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:paragraph --> <strong>1. Caching Results</strong> <!-- /wp:paragraph --> <!-- wp:paragraph --> Cache results to improve response times for repeated requests. <!-- /wp:paragraph --> <!-- wp:paragraph --> <strong>Using Redis for Caching:</strong> <!-- /wp:paragraph --> <!-- wp:preformatted --> <pre class="wp-block-preformatted">pythonCopy code<code>import redis redis_client = redis.StrictRedis(host="localhost", port=6379, decode_responses=True) @app.post("/predict") def predict(input_data: SequenceInput): sequence = input_data.sequence.upper() cache_key = f"embedding:{sequence}" # Check cache cached_embedding = redis_client.get(cache_key) if cached_embedding: return {"sequence": sequence, "embedding": eval(cached_embedding)} # Run prediction batch_labels, batch_strs, batch_tokens = batch_converter([("input", sequence)]) with torch.no_grad(): result = model(batch_tokens, repr_layers=[33]) embedding = result["representations"][33][0].tolist() # Store in cache redis_client.set(cache_key, str(embedding)) return {"sequence": sequence, "embedding": embedding} </code></pre> <!-- /wp:preformatted --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:paragraph --> <strong>2. Load Testing</strong> <!-- /wp:paragraph --> <!-- wp:paragraph --> Use tools like <strong>Apache Benchmark</strong> or <strong>k6</strong> to evaluate API performance. <!-- /wp:paragraph --> <!-- wp:paragraph --> <strong>Apache Benchmark Example:</strong> <!-- /wp:paragraph --> <!-- wp:preformatted --> <pre class="wp-block-preformatted">bashCopy code<code>ab -n 1000 -c 10 http://localhost:8000/healthcheck </code></pre> <!-- /wp:preformatted --> <!-- wp:paragraph --> <strong>k6 Test Script:</strong> <!-- /wp:paragraph --> <!-- wp:preformatted --> <pre class="wp-block-preformatted">javascriptCopy code<code>import http from 'k6/http'; import { sleep } from 'k6'; export default function () { http.post('http://localhost:8000/predict', JSON.stringify({ sequence: "MKTLLILAVVAAALA" }), { headers: { "Content-Type": "application/json" } }); sleep(1); } </code></pre> <!-- /wp:preformatted --> <!-- wp:paragraph --> Run the test: <!-- /wp:paragraph --> <!-- wp:preformatted --> <pre class="wp-block-preformatted">bashCopy code<code>k6 run script.js </code></pre> <!-- /wp:preformatted --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:paragraph --> This chapter provided a comprehensive guide to serving ESM3 models through APIs, covering setup, deployment, and optimization. With FastAPI, you can deliver real-time predictions efficiently and integrate ESM3 into scalable systems. The next chapter will focus on monitoring and maintaining production deployments. <!-- /wp:paragraph --> <!-- wp:heading {"level":3} --> <h3 class="wp-block-heading"><strong>7. Monitoring and Maintaining ESM3 in Production</strong></h3> <!-- /wp:heading --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:paragraph --> Running ESM3 in production requires robust monitoring and maintenance to ensure reliability, scalability, and performance. This chapter provides a detailed guide to setting up monitoring tools, defining maintenance workflows, and implementing best practices for maintaining ESM3 deployments. <!-- /wp:paragraph --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>7.1 The Importance of Monitoring in Production</strong></h4> <!-- /wp:heading --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:paragraph --> Monitoring production systems is essential for: <!-- /wp:paragraph --> <!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li><strong>Performance Tracking:</strong> Ensuring the ESM3 API handles requests within acceptable latency.</li> <!-- /wp:list-item --> <!-- wp:list-item --> <li><strong>Error Detection:</strong> Identifying and resolving issues such as crashes or prediction errors.</li> <!-- /wp:list-item --> <!-- wp:list-item --> <li><strong>Resource Optimization:</strong> Tracking GPU, CPU, and memory utilization to optimize costs.</li> <!-- /wp:list-item --></ul> <!-- /wp:list --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:paragraph --> <strong>Key Monitoring Areas for ESM3:</strong> <!-- /wp:paragraph --> <!-- wp:list {"ordered":true} --> <ol class="wp-block-list"><!-- wp:list-item --> <li><strong>System Metrics:</strong> CPU, GPU, and memory usage.</li> <!-- /wp:list-item --> <!-- wp:list-item --> <li><strong>Application Metrics:</strong> Request latency, throughput, and error rates.</li> <!-- /wp:list-item --> <!-- wp:list-item --> <li><strong>Model-Specific Metrics:</strong> Prediction confidence, batch processing times, and embedding quality.</li> <!-- /wp:list-item --></ol> <!-- /wp:list --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>7.2 Setting Up System Monitoring</strong></h4> <!-- /wp:heading --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:paragraph --> System monitoring involves tracking the hardware and infrastructure resources used by ESM3. <!-- /wp:paragraph --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:paragraph --> <strong>1. Using Prometheus and Grafana</strong> <!-- /wp:paragraph --> <!-- wp:paragraph --> Prometheus and Grafana are popular open-source tools for monitoring and visualizing metrics. <!-- /wp:paragraph --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:paragraph --> <strong>Installing Prometheus:</strong> <!-- /wp:paragraph --> <!-- wp:preformatted --> <pre class="wp-block-preformatted">bashCopy code<code>kubectl apply -f https://raw.githubusercontent.com/prometheus-operator/prometheus-operator/main/bundle.yaml </code></pre> <!-- /wp:preformatted --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:paragraph --> <strong>Configuring Prometheus for GPU Metrics:</strong> Install the NVIDIA DCGM exporter for GPU metrics: <!-- /wp:paragraph --> <!-- wp:preformatted --> <pre class="wp-block-preformatted">bashCopy code<code>kubectl apply -f https://raw.githubusercontent.com/NVIDIA/dcgm-exporter/main/deployments/k8s/dcgm-exporter.yaml </code></pre> <!-- /wp:preformatted --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:paragraph --> <strong>Setting Up Grafana:</strong> <!-- /wp:paragraph --> <!-- wp:list {"ordered":true} --> <ol class="wp-block-list"><!-- wp:list-item --> <li>Deploy Grafana:bashCopy code<code>kubectl apply -f https://raw.githubusercontent.com/grafana/helm-charts/main/charts/grafana/templates/deployment.yaml</code></li> <!-- /wp:list-item --> <!-- wp:list-item --> <li>Access Grafana UI:bashCopy code<code>kubectl port-forward svc/grafana 3000:3000</code></li> <!-- /wp:list-item --> <!-- wp:list-item --> <li>Add Prometheus as a data source and import dashboards for GPU monitoring.</li> <!-- /wp:list-item --></ol> <!-- /wp:list --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:paragraph --> <strong>Creating Dashboards:</strong> <!-- /wp:paragraph --> <!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li><strong>System Metrics Dashboard:</strong> Visualize GPU memory usage, GPU utilization, and GPU power consumption.</li> <!-- /wp:list-item --> <!-- wp:list-item --> <li><strong>Application Metrics Dashboard:</strong> Track API latency, request counts, and error rates.</li> <!-- /wp:list-item --></ul> <!-- /wp:list --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>7.3 Monitoring Application Metrics</strong></h4> <!-- /wp:heading --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:paragraph --> <strong>1. Logging with Fluentd and Elasticsearch</strong> <!-- /wp:paragraph --> <!-- wp:paragraph --> Logs provide granular details about application behavior and errors. <!-- /wp:paragraph --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:paragraph --> <strong>Setting Up Fluentd:</strong> <!-- /wp:paragraph --> <!-- wp:preformatted --> <pre class="wp-block-preformatted">bashCopy code<code>kubectl apply -f https://raw.githubusercontent.com/fluent/fluentd-kubernetes-daemonset/master/fluentd-daemonset-elasticsearch-rbac.yaml </code></pre> <!-- /wp:preformatted --> <!-- wp:paragraph --> <strong>Configuring Elasticsearch:</strong> Deploy Elasticsearch for log aggregation: <!-- /wp:paragraph --> <!-- wp:preformatted --> <pre class="wp-block-preformatted">bashCopy code<code>kubectl apply -f https://download.elastic.co/downloads/eck/2.1.0/all-in-one.yaml </code></pre> <!-- /wp:preformatted --> <!-- wp:paragraph --> <strong>Visualizing Logs in Kibana:</strong> Access logs via Kibana to analyze request patterns and troubleshoot errors. <!-- /wp:paragraph --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:paragraph --> <strong>2. Distributed Tracing with Jaeger</strong> <!-- /wp:paragraph --> <!-- wp:paragraph --> Jaeger enables tracing of individual requests across the system to identify performance bottlenecks. <!-- /wp:paragraph --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:paragraph --> <strong>Deploying Jaeger:</strong> <!-- /wp:paragraph --> <!-- wp:preformatted --> <pre class="wp-block-preformatted">bashCopy code<code>kubectl apply -f https://github.com/jaegertracing/jaeger-operator/releases/download/v1.33.0/jaeger-operator.yaml </code></pre> <!-- /wp:preformatted --> <!-- wp:paragraph --> <strong>Integrating with FastAPI:</strong> Install the <code>opentelemetry</code> library: <!-- /wp:paragraph --> <!-- wp:preformatted --> <pre class="wp-block-preformatted">bashCopy code<code>pip install opentelemetry-api opentelemetry-sdk opentelemetry-instrumentation-fastapi </code></pre> <!-- /wp:preformatted --> <!-- wp:paragraph --> Instrument the API: <!-- /wp:paragraph --> <!-- wp:preformatted --> <pre class="wp-block-preformatted">pythonCopy code<code>from opentelemetry.instrumentation.fastapi import FastAPIInstrumentor FastAPIInstrumentor.instrument_app(app) </code></pre> <!-- /wp:preformatted --> <!-- wp:paragraph --> Visualize traces in Jaeger to understand request flows and latency. <!-- /wp:paragraph --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>7.4 Monitoring Model-Specific Metrics</strong></h4> <!-- /wp:heading --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:paragraph --> <strong>1. Prediction Metrics</strong> <!-- /wp:paragraph --> <!-- wp:paragraph --> Track model-specific metrics like prediction confidence and embedding generation time. <!-- /wp:paragraph --> <!-- wp:paragraph --> <strong>Example: Logging Prediction Confidence:</strong> <!-- /wp:paragraph --> <!-- wp:preformatted --> <pre class="wp-block-preformatted">pythonCopy code<code>import logging logging.basicConfig(filename="model_metrics.log", level=logging.INFO) @app.post("/predict") def predict(input_data: SequenceInput): # (Prediction logic here) confidence = compute_confidence(result) # Hypothetical confidence function logging.info(f"Prediction confidence: {confidence}") return {"sequence": sequence, "confidence": confidence} </code></pre> <!-- /wp:preformatted --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:paragraph --> <strong>2. Embedding Quality Analysis</strong> <!-- /wp:paragraph --> <!-- wp:paragraph --> Track embedding outliers or patterns using statistical analysis. <!-- /wp:paragraph --> <!-- wp:paragraph --> <strong>Example: Analyzing Embedding Norms:</strong> <!-- /wp:paragraph --> <!-- wp:preformatted --> <pre class="wp-block-preformatted">pythonCopy code<code>import numpy as np @app.post("/predict") def predict(input_data: SequenceInput): # (Prediction logic here) embedding = result["representations"][33][0].tolist() norm = np.linalg.norm(embedding) logging.info(f"Embedding norm: {norm}") return {"sequence": sequence, "embedding_norm": norm} </code></pre> <!-- /wp:preformatted --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>7.5 Maintenance Best Practices</strong></h4> <!-- /wp:heading --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:paragraph --> <strong>1. Rolling Updates</strong> <!-- /wp:paragraph --> <!-- wp:paragraph --> Deploy updates to ESM3 models or APIs without downtime using Kubernetes. <!-- /wp:paragraph --> <!-- wp:paragraph --> <strong>Example: Rolling Update Command:</strong> <!-- /wp:paragraph --> <!-- wp:preformatted --> <pre class="wp-block-preformatted">bashCopy code<code>kubectl set image deployment/esm3-api esm3-api=mydockerhub/esm3-api:new-version </code></pre> <!-- /wp:preformatted --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:paragraph --> <strong>2. Backup and Disaster Recovery</strong> <!-- /wp:paragraph --> <!-- wp:paragraph --> Ensure backups of model artifacts, logs, and monitoring data. <!-- /wp:paragraph --> <!-- wp:paragraph --> <strong>Backing Up Model Artifacts:</strong> <!-- /wp:paragraph --> <!-- wp:preformatted --> <pre class="wp-block-preformatted">bashCopy code<code>aws s3 cp /models/esm3/ s3://my-backup-bucket/esm3-backup/ --recursive </code></pre> <!-- /wp:preformatted --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:paragraph --> <strong>3. Scheduled Retraining</strong> <!-- /wp:paragraph --> <!-- wp:paragraph --> Automate retraining workflows to keep models updated with the latest data. <!-- /wp:paragraph --> <!-- wp:paragraph --> <strong>Airflow DAG for Retraining:</strong> <!-- /wp:paragraph --> <!-- wp:preformatted --> <pre class="wp-block-preformatted">pythonCopy code<code>from airflow import DAG from airflow.operators.bash_operator import BashOperator from datetime import datetime default_args = {"start_date": datetime(2023, 1, 1)} with DAG("esm3_retraining", default_args=default_args, schedule_interval="@monthly") as dag: retrain = BashOperator( task_id="retrain_model", bash_command="python retrain_esm3.py" ) </code></pre> <!-- /wp:preformatted --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>7.6 Practical Example: End-to-End Monitoring</strong></h4> <!-- /wp:heading --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:paragraph --> <strong>Scenario:</strong> A bioinformatics lab needs to monitor an ESM3 deployment for prediction accuracy, system utilization, and application errors. <!-- /wp:paragraph --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:paragraph --> <strong>Steps:</strong> <!-- /wp:paragraph --> <!-- wp:list {"ordered":true} --> <ol class="wp-block-list"><!-- wp:list-item --> <li><strong>Set Up Prometheus and Grafana:</strong><!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Monitor GPU metrics and API latency.</li> <!-- /wp:list-item --> <!-- wp:list-item --> <li>Create alerts for high GPU usage.</li> <!-- /wp:list-item --></ul> <!-- /wp:list --></li> <!-- /wp:list-item --> <!-- wp:list-item --> <li><strong>Configure Logging:</strong><!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Aggregate logs using Fluentd and Elasticsearch.</li> <!-- /wp:list-item --> <!-- wp:list-item --> <li>Analyze error trends in Kibana.</li> <!-- /wp:list-item --></ul> <!-- /wp:list --></li> <!-- /wp:list-item --> <!-- wp:list-item --> <li><strong>Enable Distributed Tracing:</strong><!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Trace prediction latency with Jaeger.</li> <!-- /wp:list-item --> <!-- wp:list-item --> <li>Identify bottlenecks in batch prediction workflows.</li> <!-- /wp:list-item --></ul> <!-- /wp:list --></li> <!-- /wp:list-item --> <!-- wp:list-item --> <li><strong>Track Prediction Confidence:</strong><!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Log confidence scores and visualize trends in Grafana.</li> <!-- /wp:list-item --></ul> <!-- /wp:list --></li> <!-- /wp:list-item --> <!-- wp:list-item --> <li><strong>Automate Maintenance:</strong><!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Use Airflow to schedule model retraining.</li> <!-- /wp:list-item --> <!-- wp:list-item --> <li>Implement Kubernetes rolling updates for new deployments.</li> <!-- /wp:list-item --></ul> <!-- /wp:list --></li> <!-- /wp:list-item --></ol> <!-- /wp:list --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:paragraph --> <strong>Example Workflow:</strong> <!-- /wp:paragraph --> <!-- wp:preformatted --> <pre class="wp-block-preformatted">bashCopy code<code># Deploy Prometheus and Grafana kubectl apply -f prometheus-operator.yaml kubectl apply -f grafana-deployment.yaml # Start the FastAPI server with tracing enabled uvicorn main:app --host 0.0.0.0 --port 8000 # Analyze logs in Kibana kubectl logs <esm3-api-pod-name> | grep ERROR </code></pre> <!-- /wp:preformatted --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:paragraph --> This chapter provided a comprehensive guide to monitoring and maintaining ESM3 in production, covering system metrics, application logs, and model-specific insights. With tools like Prometheus, Grafana, and Jaeger, you can ensure your deployment is robust, performant, and reliable. The next chapter will focus on optimizing costs and scaling ESM3 for large-scale production use cases. <!-- /wp:paragraph --> <!-- wp:heading {"level":3} --> <h3 class="wp-block-heading"><strong>8. Optimizing Costs and Scaling ESM3 for Large-Scale Production</strong></h3> <!-- /wp:heading --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:paragraph --> Deploying ESM3 in production environments often involves high computational demands. This chapter explores strategies to optimize costs while scaling for large workloads. It includes practical approaches to resource allocation, model optimization, and leveraging cloud solutions. <!-- /wp:paragraph --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>8.1 Understanding Cost Drivers</strong></h4> <!-- /wp:heading --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:paragraph --> Before optimizing costs, it is essential to identify the key drivers of expenses in ESM3 deployments. <!-- /wp:paragraph --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:paragraph --> <strong>Key Cost Drivers:</strong> <!-- /wp:paragraph --> <!-- wp:list {"ordered":true} --> <ol class="wp-block-list"><!-- wp:list-item --> <li><strong>Infrastructure:</strong><!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>GPU instances for inference.</li> <!-- /wp:list-item --> <!-- wp:list-item --> <li>Storage for model artifacts and datasets.</li> <!-- /wp:list-item --></ul> <!-- /wp:list --></li> <!-- /wp:list-item --> <!-- wp:list-item --> <li><strong>Workload Size:</strong><!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Batch sizes and frequency of predictions.</li> <!-- /wp:list-item --> <!-- wp:list-item --> <li>Volume of sequences processed.</li> <!-- /wp:list-item --></ul> <!-- /wp:list --></li> <!-- /wp:list-item --> <!-- wp:list-item --> <li><strong>Inefficient Resource Utilization:</strong><!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Overprovisioning compute resources.</li> <!-- /wp:list-item --> <!-- wp:list-item --> <li>Underutilized GPU or CPU resources.</li> <!-- /wp:list-item --></ul> <!-- /wp:list --></li> <!-- /wp:list-item --> <!-- wp:list-item --> <li><strong>Scaling Overheads:</strong><!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Inefficient scaling configurations leading to unnecessary resource consumption.</li> <!-- /wp:list-item --></ul> <!-- /wp:list --></li> <!-- /wp:list-item --></ol> <!-- /wp:list --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:paragraph --> <strong>Cost Analysis Example:</strong> For a typical ESM3 deployment processing 1,000 predictions per hour: <!-- /wp:paragraph --> <!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li><strong>GPU Cost:</strong>](https://esm3academy.com/wp-content/ql-cache/quicklatex.com-6d170cf632a39da5860a270a94265825_l3.png) 3 per hour (AWS g4dn.xlarge).
3 per hour (AWS g4dn.xlarge).
Storage Cost: 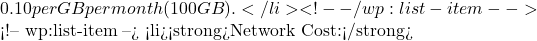 0.09 per GB of data transfer (50 GB/month).
0.09 per GB of data transfer (50 GB/month).
By optimizing these components, significant savings can be achieved.
8.2 Resource Optimization
1. Right-Sizing Compute Instances
Choose compute resources that align with workload requirements.
Example Instance Selection:
- For Small Workloads: AWS g4dn.xlarge (1 GPU, 16 GB RAM).
- For Medium Workloads: AWS p3.2xlarge (1 V100 GPU, 64 GB RAM).
- For Large Workloads: AWS p3.8xlarge (4 V100 GPUs, 256 GB RAM).
Best Practices:
- Monitor utilization with tools like Prometheus to prevent overprovisioning.
- Use auto-scaling groups to dynamically adjust resources.
2. Using Spot Instances
Spot instances offer significant cost savings compared to on-demand instances but are subject to interruptions.
Example Configuration:
bashCopy codeaws ec2 run-instances --instance-type g4dn.xlarge --spot-price "0.10" --image-id ami-12345678
Use Kubernetes with spot instance support:
yamlCopy codeapiVersion: apps/v1
kind: Deployment
metadata:
name: esm3-deployment
spec:
replicas: 2
template:
spec:
nodeSelector:
"lifecycle": "spot"
3. Efficient Resource Allocation
Control GPU and CPU allocation for containers running ESM3.
Example: Kubernetes Resource Requests and Limits
yamlCopy coderesources:
requests:
memory: "8Gi"
cpu: "2"
nvidia.com/gpu: "1"
limits:
memory: "16Gi"
cpu: "4"
nvidia.com/gpu: "1"
4. Batch Processing
Process multiple sequences in a single batch to maximize GPU utilization.
Batch Prediction Script:
pythonCopy codedef batch_process(sequences, batch_size):
for i in range(0, len(sequences), batch_size):
batch = sequences[i:i + batch_size]
yield batch
sequences = ["MKTLLILAVVAAALA", "ACDEFGHIKLMNPQRSTVWY", "MKTLLIMVVVAAGLA"]
for batch in batch_process(sequences, batch_size=2):
# Run ESM3 predictions on the batch
print(batch)
8.3 Model Optimization
1. Quantization
Quantization reduces the precision of model weights and activations to lower computational requirements.
Using PyTorch Quantization:
pythonCopy codeimport torch
from esm import pretrained
model, alphabet = pretrained.esm3_t36_3B_UR50D()
# Quantize the model
model = torch.quantization.quantize_dynamic(
model, {torch.nn.Linear}, dtype=torch.qint8
)
Benefits:
- Reduces model size and inference latency.
- Slight trade-off in accuracy.
2. Model Pruning
Pruning removes less critical parameters from the model to reduce size and improve efficiency.
Example: Pruning with PyTorch:
pythonCopy codefrom torch.nn.utils import prune
for name, module in model.named_modules():
if isinstance(module, torch.nn.Linear):
prune.l1_unstructured(module, name="weight", amount=0.2)
3. Caching Embeddings
For repetitive sequences, cache embeddings to avoid redundant computation.
Using Redis for Caching:
pythonCopy codeimport redis
redis_client = redis.StrictRedis(host="localhost", port=6379)
def get_embedding(sequence):
cached_embedding = redis_client.get(sequence)
if cached_embedding:
return eval(cached_embedding)
else:
# Compute embedding with ESM3
embedding = model.predict(sequence)
redis_client.set(sequence, str(embedding))
return embedding
8.4 Scaling ESM3
1. Horizontal Scaling
Scale ESM3 deployments horizontally by adding more instances.
Kubernetes Horizontal Pod Autoscaler:
bashCopy codekubectl autoscale deployment esm3-deployment --cpu-percent=70 --min=2 --max=10
2. Vertical Scaling
Increase resources per instance to handle larger workloads.
Update Kubernetes deployment:
yamlCopy coderesources:
requests:
memory: "16Gi"
cpu: "8"
3. Hybrid Scaling
Combine spot and on-demand instances for cost-effective scaling.
Kubernetes Node Affinity:
yamlCopy codespec:
nodeSelector:
"lifecycle": "spot"
tolerations:
- key: "spot"
operator: "Exists"
effect: "NoSchedule"
4. Load Balancing
Distribute incoming traffic evenly across instances.
Using Kubernetes LoadBalancer:
yamlCopy codeapiVersion: v1
kind: Service
metadata:
name: esm3-loadbalancer
spec:
type: LoadBalancer
selector:
app: esm3
ports:
- protocol: TCP
port: 80
targetPort: 8000
8.5 Practical Example: Cost-Effective ESM3 Deployment
Scenario: A research institute processes 10,000 protein sequences daily using ESM3. The goal is to reduce costs by 30%.
Steps:
- Use AWS spot instances for inference workloads.
- Implement batch processing to maximize GPU utilization.
- Quantize the ESM3 model to reduce latency.
- Deploy Kubernetes autoscaling with resource requests optimized for cost.
End-to-End Workflow:
bashCopy code# Launch spot instances
aws ec2 run-instances --instance-type g4dn.xlarge --spot-price "0.10" --image-id ami-12345678
# Deploy ESM3 on Kubernetes
kubectl apply -f esm3-deployment.yaml
kubectl apply -f esm3-loadbalancer.yaml
# Optimize model
python optimize_model.py
# Enable autoscaling
kubectl autoscale deployment esm3-deployment --cpu-percent=80 --min=1 --max=5
This chapter provided a detailed guide to optimizing costs and scaling ESM3 deployments. From resource allocation and model optimization to leveraging cloud solutions, these strategies ensure efficient, cost-effective operations. The next chapter will focus on ensuring security and compliance for production deployments.
9. Ensuring Security and Compliance in ESM3 Deployments
Deploying ESM3 in production environments requires robust security measures to protect sensitive data and ensure compliance with industry standards. This chapter provides a comprehensive guide to implementing security best practices, managing data protection, and ensuring compliance with regulatory frameworks.
9.1 Importance of Security and Compliance
Security and compliance are critical for:
- Data Protection: Safeguarding sensitive biological data and research outputs.
- Regulatory Compliance: Meeting standards such as GDPR, HIPAA, and CCPA for data privacy.
- System Integrity: Preventing unauthorized access, data breaches, and malware attacks.
Key Security Challenges:
- Protecting sensitive protein sequence data during transmission and storage.
- Managing access to ESM3 APIs and models.
- Ensuring auditability and traceability of data processing activities.
9.2 Securing Data Transmission
Data transmitted between clients and servers must be encrypted to prevent interception by unauthorized parties.
1. Enabling HTTPS
Use TLS (Transport Layer Security) to encrypt API communications.
Generating TLS Certificates with Let's Encrypt:
bashCopy codesudo apt install certbot
sudo certbot certonly --standalone -d example.com
Update the server configuration to use HTTPS:
bashCopy codeuvicorn main:app --host 0.0.0.0 --port 443 --ssl-keyfile /etc/letsencrypt/live/example.com/privkey.pem --ssl-certfile /etc/letsencrypt/live/example.com/fullchain.pem
2. Encrypting API Payloads
Encrypt sensitive payloads before sending them to the API.
Example: Encrypting Payloads with Fernet (Python):
pythonCopy codefrom cryptography.fernet import Fernet
# Generate a key
key = Fernet.generate_key()
cipher = Fernet(key)
# Encrypt a message
message = "MKTLLILAVVAAALA"
encrypted_message = cipher.encrypt(message.encode())
# Decrypt the message
decrypted_message = cipher.decrypt(encrypted_message).decode()
print(decrypted_message)
9.3 Securing Data Storage
Store sensitive data such as ESM3 outputs and logs in encrypted formats to protect against unauthorized access.
1. Encrypting Databases
Use database-level encryption for storing sensitive information.
Example: Encrypting SQLite Data:
pythonCopy codefrom cryptography.fernet import Fernet
import sqlite3
# Generate an encryption key
key = Fernet.generate_key()
cipher = Fernet(key)
# Encrypt data before storing it
sequence = "MKTLLILAVVAAALA"
encrypted_sequence = cipher.encrypt(sequence.encode())
conn = sqlite3.connect("secure_esm3.db")
cursor = conn.cursor()
cursor.execute("CREATE TABLE IF NOT EXISTS predictions (id INTEGER PRIMARY KEY, sequence BLOB)")
cursor.execute("INSERT INTO predictions (sequence) VALUES (?)", (encrypted_sequence,))
conn.commit()
2. Encrypting File Storage
Encrypt files containing sensitive data before saving them to disk.
Example: Encrypting Output Files:
pythonCopy codewith open("esm3_output.json", "rb") as file:
encrypted_data = cipher.encrypt(file.read())
with open("encrypted_esm3_output.json", "wb") as encrypted_file:
encrypted_file.write(encrypted_data)
9.4 Managing Access Control
Restrict access to the ESM3 API and data to authorized users and applications.
1. Implementing API Authentication
Use OAuth 2.0 or API keys for authentication.
Example: Using FastAPI with API Keys:
pythonCopy codefrom fastapi import FastAPI, HTTPException, Depends
app = FastAPI()
API_KEY = "your-secure-api-key"
def verify_api_key(api_key: str = Depends()):
if api_key != API_KEY:
raise HTTPException(status_code=403, detail="Invalid API Key")
@app.get("/predict", dependencies=[Depends(verify_api_key)])
def predict(sequence: str):
return {"message": f"Processing sequence: {sequence}"}
2. Role-Based Access Control (RBAC)
Implement RBAC to define granular permissions for different user roles.
Example: Defining Roles:
pythonCopy coderoles = {
"admin": {"access": ["read", "write", "delete"]},
"user": {"access": ["read"]},
}
def has_permission(user_role, action):
return action in roles.get(user_role, {}).get("access", [])
# Usage
if not has_permission("user", "write"):
print("Permission denied")
9.5 Logging and Auditing
Logging and auditing provide traceability for all API interactions and system activities.
1. Logging API Requests
Log all incoming requests and their responses.
Example: Logging Middleware in FastAPI:
pythonCopy codefrom fastapi import Request
import logging
logging.basicConfig(filename="esm3_api.log", level=logging.INFO)
@app.middleware("http")
async def log_requests(request: Request, call_next):
response = await call_next(request)
logging.info(f"{request.method} {request.url} - {response.status_code}")
return response
2. Audit Trails
Maintain an immutable record of sensitive actions for compliance.
Example: Storing Audit Logs:
pythonCopy codeaudit_logs = []
def log_action(user, action):
audit_logs.append({"user": user, "action": action, "timestamp": datetime.now()})
log_action("admin", "deleted_prediction")
9.6 Ensuring Compliance
Compliance with regulations is mandatory when processing sensitive data.
1. GDPR Compliance
Under GDPR, ensure:
- User Consent: Collect explicit consent for data usage.
- Right to Access/Erasure: Allow users to access and delete their data.
Example: Adding Consent to API Usage:
pythonCopy code@app.post("/consent")
def store_user_consent(user_id: str, consent: bool):
# Store consent in the database
return {"message": "Consent recorded"}
2. HIPAA Compliance
For healthcare-related data:
- Use encrypted data transmission and storage.
- Implement strict access controls and audit logs.
3. Periodic Compliance Audits
Use tools like Nessus or OpenSCAP to automate compliance checks.
Example: Running OpenSCAP:
bashCopy codeoscap xccdf eval --profile HIPAA /path/to/profile.xml
9.7 Practical Example: End-to-End Security Setup
Scenario: A biotech company deploys ESM3 to process sensitive protein data for drug discovery and needs to secure its API and storage.
Steps:
- Enable HTTPS:
- Use TLS to encrypt communications.
- Encrypt Data:
- Encrypt sensitive outputs before saving them.
- Restrict Access:
- Use API keys for authentication.
- Log and Audit:
- Implement logging middleware for all API requests.
- Compliance Checks:
- Automate GDPR and HIPAA compliance validation.
Complete Workflow:
bashCopy code# 1. Secure the API
uvicorn main:app --ssl-keyfile /path/to/privkey.pem --ssl-certfile /path/to/fullchain.pem
# 2. Set up logging
tail -f esm3_api.log
# 3. Verify compliance
oscap xccdf eval --profile GDPR /path/to/profile.xml
This chapter detailed how to secure ESM3 deployments by encrypting data, managing access, implementing logging and auditing, and ensuring compliance with regulations. With these practices, you can safeguard sensitive data and build trust with users and stakeholders. The next chapter will focus on integrating ESM3 into enterprise ecosystems for seamless adoption.
10. Integrating ESM3 into Enterprise Ecosystems
Deploying ESM3 into an enterprise ecosystem requires integration with existing tools, workflows, and platforms to ensure smooth adoption and maximize utility. This chapter explores strategies, architectures, and practical examples for seamlessly integrating ESM3 with enterprise applications.
10.1 The Enterprise Ecosystem: Key Components
An enterprise ecosystem typically includes:
- Data Management Systems: Databases, data lakes, and data warehouses.
- Business Applications: CRMs, ERPs, and custom applications.
- Collaboration Tools: Platforms like Slack, Microsoft Teams, and JIRA.
- AI/ML Platforms: Pre-existing infrastructure for training, deployment, and monitoring.
Integration Goals for ESM3:
- Enable interoperability with existing systems.
- Support workflows such as automated predictions, notifications, and reporting.
- Ensure scalability and maintainability.
10.2 Designing Integration Architectures
1. Service-Oriented Architecture (SOA)
Deploy ESM3 as a standalone service accessible via APIs. Other enterprise systems can call this API for predictions.
Architecture Example:
Component Description ESM3 API Handles prediction requests. Database Stores sequences and predictions. Orchestration Uses workflows to trigger the API (e.g., Airflow). Notification System Sends alerts based on predictions.
Example Workflow:
- A new sequence is added to the database.
- A trigger invokes the ESM3 API for prediction.
- Results are stored, and alerts are sent via Slack.
2. Event-Driven Architecture
Integrate ESM3 using event streams for real-time processing.
Architecture Example:
Component Description Message Queue Kafka or RabbitMQ for event-driven workflows. ESM3 Service Consumes events, processes sequences, and outputs results. Downstream Systems Applications that consume results.
Example Workflow:
- A Kafka topic publishes new protein sequences.
- ESM3 consumes the topic, processes sequences, and writes predictions to a results topic.
3. Microservices Architecture
Integrate ESM3 as a microservice, leveraging container orchestration for scalability.
Example Deployment with Kubernetes:
yamlCopy codeapiVersion: apps/v1
kind: Deployment
metadata:
name: esm3-service
spec:
replicas: 3
selector:
matchLabels:
app: esm3
template:
metadata:
labels:
app: esm3
spec:
containers:
- name: esm3-api
image: esm3-api:latest
ports:
- containerPort: 8000
10.3 Integrating with Data Management Systems
1. Databases
Integrate ESM3 with relational or NoSQL databases for storing sequences and predictions.
Example: PostgreSQL Integration
Create a table for storing ESM3 predictions:
sqlCopy codeCREATE TABLE esm3_predictions (
id SERIAL PRIMARY KEY,
sequence TEXT NOT NULL,
embedding JSONB NOT NULL,
created_at TIMESTAMP DEFAULT CURRENT_TIMESTAMP
);
Insert predictions directly from the API:
pythonCopy codeimport psycopg2
conn = psycopg2.connect(
dbname="enterprise_db", user="admin", password="password", host="localhost"
)
cursor = conn.cursor()
sequence = "MKTLLILAVVAAALA"
embedding = {"layer_33": [0.1, 0.2, 0.3]}
cursor.execute(
"INSERT INTO esm3_predictions (sequence, embedding) VALUES (%s, %s)",
(sequence, json.dumps(embedding)),
)
conn.commit()
2. Data Lakes
For large-scale predictions, store outputs in data lakes for further analysis.
Example: Storing Outputs in AWS S3
pythonCopy codeimport boto3
s3 = boto3.client("s3")
sequence = "MKTLLILAVVAAALA"
embedding = {"layer_33": [0.1, 0.2, 0.3]}
s3.put_object(
Bucket="esm3-predictions",
Key="predictions/sequence_1.json",
Body=json.dumps({"sequence": sequence, "embedding": embedding}),
)
3. ETL Pipelines
Build ETL pipelines to preprocess input data, invoke ESM3, and store outputs.
Example: Using Apache Airflow
pythonCopy codefrom airflow import DAG
from airflow.operators.python_operator import PythonOperator
from datetime import datetime
def run_esm3_prediction():
# Example: Invoke ESM3 API
pass
default_args = {"start_date": datetime(2023, 1, 1)}
dag = DAG("esm3_etl", default_args=default_args, schedule_interval="0 * * * *")
predict_task = PythonOperator(
task_id="predict_sequence",
python_callable=run_esm3_prediction,
dag=dag,
)
10.4 Integrating with Business Applications
1. CRM Systems
Enrich CRM data with ESM3 predictions, such as drug development insights.
Example: Salesforce Integration Use Salesforce’s REST API to update records with ESM3 outputs:
pythonCopy codeimport requests
url = "https://your-instance.salesforce.com/services/data/v53.0/sobjects/CustomObject__c"
headers = {"Authorization": "Bearer YOUR_ACCESS_TOKEN"}
data = {
"Name": "Protein Analysis",
"ESM3_Result__c": "Prediction details here",
}
response = requests.post(url, json=data, headers=headers)
2. Collaboration Tools
Send ESM3 predictions directly to collaboration tools like Slack or Microsoft Teams.
Example: Slack Notification
pythonCopy codeimport requests
slack_webhook_url = "https://hooks.slack.com/services/T00000000/B00000000/XXXXXXXXXXXXXXXXXXXXXXXX"
message = {
"text": "New ESM3 prediction completed for sequence MKTLLILAVVAAALA."
}
requests.post(slack_webhook_url, json=message)
3. Custom Applications
Embed ESM3 predictions into dashboards or reports.
Example: Embedding Predictions in Dash
pythonCopy codeimport dash
from dash import dcc, html
app = dash.Dash(__name__)
predictions = {"sequence": "MKTLLILAVVAAALA", "confidence": 0.95}
app.layout = html.Div([
html.H1("ESM3 Predictions"),
html.Div(f"Sequence: {predictions['sequence']}"),
html.Div(f"Confidence: {predictions['confidence']}")
])
if __name__ == "__main__":
app.run_server(debug=True)
10.5 Practical Example: End-to-End Integration
Scenario: A pharmaceutical company integrates ESM3 predictions into its enterprise workflow for drug discovery.
Workflow Steps:
- Data Input: Sequences are uploaded to a database.
- Prediction Trigger: A pipeline triggers ESM3 predictions.
- Result Storage: Predictions are stored in S3 for analysis.
- Notifications: Results are sent to Slack for the research team.
- Reporting: Results are visualized in a custom dashboard.
Implementation:
bashCopy code# 1. Deploy ESM3 API
uvicorn main:app --host 0.0.0.0 --port 8000
# 2. Trigger predictions using Airflow
airflow dags trigger esm3_etl
# 3. Store outputs in S3
python store_predictions.py
# 4. Notify team on Slack
python send_slack_notification.py
# 5. Visualize results
python run_dash_dashboard.py
This chapter outlined practical approaches for integrating ESM3 into enterprise ecosystems, including architecture design, data system integration, and application embedding. By following these methods, enterprises can unlock the full potential of ESM3 in their workflows. The next chapter will explore use cases and applications of ESM3 in various industries.
11. Use Cases and Applications of ESM3 in Various Industries
Evolutionary Scale Modeling 3 (ESM3) is a transformative tool for protein sequence analysis and structural predictions, with applications spanning multiple industries. This chapter explores practical use cases, implementation strategies, and examples to demonstrate how ESM3 can be applied effectively across sectors such as biotechnology, pharmaceuticals, agriculture, and more.
11.1 Applications in Biotechnology
Biotechnology relies on protein analysis to innovate in areas like enzyme engineering, synthetic biology, and molecular diagnostics. ESM3 offers precise sequence predictions and embeddings that can accelerate these processes.
1. Enzyme Engineering
Use Case: Optimize enzyme efficiency for industrial applications like biofuels or pharmaceuticals.
Workflow:
- Input: Provide sequences of target enzymes to ESM3.
- Output: Predict conserved regions and potential mutation sites.
- Optimization: Use predictions to guide mutagenesis experiments.
Example: Identifying Key Residues for Mutation
pythonCopy codesequence = "MKTLLILAVVAAALA"
esm3_predictions = esm3_api.predict(sequence)
# Analyze conserved regions
conserved_regions = [
i for i, prob in enumerate(esm3_predictions["token_probabilities"]) if prob > 0.9
]
print(f"Highly conserved residues: {conserved_regions}")
Result: Identify residues critical to enzyme function, enabling targeted improvements.
2. Molecular Diagnostics
Use Case: Detect biomarkers or mutations associated with diseases.
Workflow:
- Input: Provide patient-derived protein sequences.
- Output: Use ESM3 to predict structural changes due to mutations.
- Clinical Interpretation: Combine predictions with patient data for diagnostics.
Example: Predicting Mutation Effects
pythonCopy codemutant_sequence = "MKTLLILVVAAALA"
wild_type_sequence = "MKTLLILAVVAAALA"
wild_type_structure = esm3_api.predict(wild_type_sequence)["3D_structure"]
mutant_structure = esm3_api.predict(mutant_sequence)["3D_structure"]
# Compare structures
compare_structures(wild_type_structure, mutant_structure)
11.2 Applications in Pharmaceuticals
ESM3 is a powerful tool for drug discovery and development, enabling researchers to identify drug targets, optimize protein-ligand interactions, and explore potential side effects.
1. Drug Target Identification
Use Case: Discover novel drug targets by analyzing conserved regions in protein families.
Workflow:
- Input: Analyze sequences across a protein family.
- Output: Identify conserved regions and potential binding sites.
- Validation: Use structural predictions to validate target feasibility.
Example: Identifying a Drug Binding Site
pythonCopy codesequence = "MKTLLILAVVAAALA"
predictions = esm3_api.predict(sequence)
# Highlight potential binding sites
binding_sites = [
i for i, score in enumerate(predictions["token_probabilities"]) if score > 0.85
]
print(f"Potential binding sites: {binding_sites}")
2. Protein-Ligand Docking
Use Case: Model protein-ligand interactions to optimize drug candidates.
Workflow:
- Input: Provide ESM3-predicted structures for docking simulations.
- Output: Model protein-ligand binding and calculate binding affinities.
- Optimization: Modify ligands for improved binding.
Example: Docking Simulations Using PyMOL
bashCopy code# Load ESM3-predicted structure
pymol -c -d "load esm3_structure.pdb; show surface; load ligand.pdb; dock ligand, esm3_structure"
11.3 Applications in Agriculture
Protein sequence analysis plays a critical role in agricultural biotechnology, enabling advancements in crop protection, livestock health, and pest resistance.
1. Pest Resistance
Use Case: Develop pest-resistant crops by analyzing plant proteins targeted by pests.
Workflow:
- Input: Provide sequences of plant proteins susceptible to pests.
- Output: Use ESM3 to predict resistant mutations.
- Implementation: Introduce mutations through gene editing.
Example: Predicting Resistance Mutations
pythonCopy codeplant_protein = "MKTLLILAVVAAALA"
predictions = esm3_api.predict(plant_protein)
# Identify low-confidence regions (susceptible to mutations)
susceptible_regions = [
i for i, score in enumerate(predictions["token_probabilities"]) if score < 0.7
]
print(f"Regions for potential resistance mutations: {susceptible_regions}")
2. Livestock Health
Use Case: Identify protein markers for disease resistance in livestock.
Workflow:
- Input: Analyze protein sequences associated with immunity.
- Output: Predict mutations to enhance resistance.
- Implementation: Breed livestock with beneficial mutations.
Example: Enhancing Immunity
pythonCopy codesequence = "MKTLLILAVVAAALA"
embedding = esm3_api.predict(sequence)["embedding"]
# Cluster similar immune-related proteins
clusters = cluster_embeddings(embedding, method="kmeans", num_clusters=3)
11.4 Applications in Academia and Research
ESM3 supports academic research by enabling large-scale protein analyses, structural studies, and evolutionary investigations.
1. Evolutionary Studies
Use Case: Explore evolutionary relationships between proteins across species.
Workflow:
- Input: Provide protein sequences from multiple species.
- Output: Generate embeddings and cluster proteins based on similarity.
- Analysis: Identify conserved domains and phylogenetic relationships.
Example: Clustering Protein Families
pythonCopy codesequences = ["MKTLLILAVVAAALA", "ACDEFGHIKLMNPQRS", "MKTLLIMVVVAAGLA"]
embeddings = [esm3_api.predict(seq)["embedding"] for seq in sequences]
# Dimensionality reduction and clustering
reduced_embeddings = pca_reduce(embeddings)
clusters = kmeans_clustering(reduced_embeddings, n_clusters=2)
2. Structural Studies
Use Case: Investigate protein folding and stability using ESM3’s 3D predictions.
Workflow:
- Input: Provide sequences of proteins with unknown structures.
- Output: Predict 3D structures and assess folding stability.
- Analysis: Use predictions to hypothesize protein function.
Example: Visualizing 3D Structures with Py3Dmol
pythonCopy codeimport py3Dmol
pdb_data = esm3_api.predict(sequence)["3D_structure"]
viewer = py3Dmol.view()
viewer.addModel(pdb_data, "pdb")
viewer.setStyle({"cartoon": {"color": "spectrum"}})
viewer.zoomTo()
viewer.show()
11.5 Practical Example: Cross-Industry Workflow
Scenario: An interdisciplinary project seeks to identify proteins for drug discovery, crop protection, and evolutionary studies. The team needs to integrate ESM3 predictions across these domains.
Steps:
- Input Data: Collect sequences from pharmaceutical, agricultural, and academic datasets.
- Prediction: Use ESM3 for structural and functional predictions.
- Analysis: Group predictions by application and visualize results.
- Implementation: Use results for drug target validation, crop genetic modifications, and evolutionary analysis.
End-to-End Workflow:
pythonCopy code# Collect sequences
sequences = ["MKTLLILAVVAAALA", "ACDEFGHIKLMNPQRS", "MKTLLIMVVVAAGLA"]
# Predict for all sequences
results = [esm3_api.predict(seq) for seq in sequences]
# Analyze results
for res in results:
analyze_prediction(res)
This chapter provided a comprehensive overview of how ESM3 can be applied across various industries, including biotechnology, pharmaceuticals, agriculture, and academia. With practical workflows and examples, you can leverage ESM3’s capabilities to drive innovation in your field. The next chapter will explore future developments and trends in ESM3 applications.
12. Future Directions and Trends in ESM3 Deployments
As machine learning and bioinformatics continue to evolve, ESM3 stands poised to shape the future of protein analysis and structural biology. This chapter explores emerging trends, potential advancements, and new areas of application for ESM3 in both research and industry.
12.1 Advances in Model Optimization
The capabilities of ESM3 can be significantly enhanced through ongoing optimization techniques, ensuring better performance and broader accessibility.
1. Multi-Modal Integration
Future iterations of ESM models may incorporate additional data types, such as RNA sequences, chemical properties, or protein-ligand interactions. Integrating these modalities can provide a holistic view of biomolecular systems.
Example: Integrating RNA and Protein Predictions
- Use RNA sequences as supplementary input to predict protein function more accurately.
- Combine protein embeddings with ligand structures for drug discovery.
Hypothetical Workflow:
pythonCopy codeprotein_sequence = "MKTLLILAVVAAALA"
rna_sequence = "AUGAUGCUCUGAAUUA"
# Process RNA and protein sequences with multimodal ESM
rna_embedding = rna_model.predict(rna_sequence)
protein_embedding = esm3_model.predict(protein_sequence)
# Combine embeddings for downstream analysis
combined_embedding = combine_embeddings(rna_embedding, protein_embedding)
2. Federated Learning
To protect sensitive data, federated learning could allow ESM3 to be trained collaboratively across institutions without sharing raw data. This ensures privacy and enhances the model with diverse datasets.
Example: Training Federated Models
pythonCopy codefrom federated_learning import FederatedTrainer
trainer = FederatedTrainer(models=[esm3_model], data_sources=["lab1", "lab2", "lab3"])
trainer.train()
3. Improved Interpretability
Interpretable AI methods are expected to make ESM3 predictions more transparent. By visualizing attention mechanisms or highlighting critical residues, researchers can better understand the model's decisions.
Example: Visualizing Attention in ESM3
pythonCopy codeimport matplotlib.pyplot as plt
attention_weights = esm3_model.get_attention(sequence)
plt.imshow(attention_weights, cmap="hot")
plt.title("Attention Map for Sequence")
plt.show()
12.2 Enhanced Deployment Mechanisms
Deployment strategies will evolve to make ESM3 more accessible, scalable, and efficient for a wider audience.
1. Serverless Deployments
Serverless frameworks such as AWS Lambda or Google Cloud Functions enable cost-effective and scalable deployments.
Example: Deploying ESM3 with AWS Lambda
bashCopy code# Package the ESM3 model
zip -r esm3_lambda.zip esm3_model/
# Deploy to Lambda
aws lambda create-function \
--function-name ESM3Prediction \
--runtime python3.8 \
--handler handler.predict \
--zip-file fileb://esm3_lambda.zip
2. Edge Computing
Bringing ESM3 to edge devices, such as portable genomic analyzers or field equipment, could empower real-time protein predictions in remote locations.
Example: Using TensorFlow Lite for Edge Deployment
pythonCopy codeimport tensorflow as tf
# Convert ESM3 model to TensorFlow Lite format
converter = tf.lite.TFLiteConverter.from_saved_model("esm3_model/")
tflite_model = converter.convert()
# Save the model for deployment
with open("esm3_model.tflite", "wb") as f:
f.write(tflite_model)
3. Cloud-Native Pipelines
Cloud-native technologies like Kubernetes and serverless databases will streamline large-scale ESM3 operations.
Example: Using Kubernetes for Workflow Automation
yamlCopy codeapiVersion: batch/v1
kind: Job
metadata:
name: esm3-prediction-job
spec:
template:
spec:
containers:
- name: esm3
image: esm3-predictor:latest
command: ["python", "predict.py", "--sequence", "MKTLLILAVVAAALA"]
restartPolicy: Never
12.3 Expanding Applications
New applications for ESM3 will emerge as the field of computational biology grows and interdisciplinary approaches gain momentum.
1. Synthetic Biology
ESM3 can be used to design synthetic proteins with desired properties, such as enhanced stability, functionality, or specificity.
Example: Designing a Synthetic Enzyme
pythonCopy codefrom esm3_optimizer import EnzymeDesigner
designer = EnzymeDesigner(model=esm3_model)
optimized_sequence = designer.optimize("MKTLLILAVVAAALA", target_function="stability")
print(f"Optimized sequence: {optimized_sequence}")
2. Personalized Medicine
As precision medicine evolves, ESM3 could help tailor treatments based on individual protein variations, predicting the efficacy of therapies.
Example: Predicting Drug Resistance
pythonCopy codemutated_protein = "MKTLLILVLVAAALA"
# Use ESM3 to predict mutation impact
prediction = esm3_model.predict(mutated_protein)
print(f"Drug resistance likelihood: {prediction['resistance_score']}")
3. Environmental Science
Incorporating ESM3 into environmental research can aid in understanding microbial ecosystems or biodegradation pathways.
Example: Exploring Microbial Protein Functions
pythonCopy codemicrobial_sequence = "ACDEFGHIKLMNPQRSTVWY"
# Predict function of microbial proteins
function_prediction = esm3_model.predict_function(microbial_sequence)
print(f"Predicted function: {function_prediction}")
12.4 Challenges and Considerations
As ESM3 applications expand, addressing key challenges will be essential for sustainable growth.
1. Ethical Considerations
Responsible usage of ESM3 requires addressing concerns such as:
- Misuse of synthetic biology capabilities.
- Fair access to ESM3 technologies for low-resource settings.
2. Data Privacy
Ensuring the secure processing of sensitive biological and medical data remains critical.
3. Computational Resources
The increasing complexity of models demands more efficient infrastructure and algorithms to minimize energy consumption.
12.5 Collaborative Innovations
The future of ESM3 lies in collaborative innovation across industries, research institutions, and regulatory bodies.
1. Open-Source Ecosystems
Expanding open-source libraries for ESM3 will promote accessibility and accelerate innovation.
2. Industry-Academia Partnerships
Collaborations between academia and industries can drive translational research, bringing ESM3 findings into real-world applications.
3. Standardization
Establishing standards for ESM3 predictions, embeddings, and annotations will enable seamless integration across platforms.
The future of ESM3 is rich with possibilities, from enhancing its capabilities through optimization techniques to expanding its reach into new industries and applications. By addressing challenges and fostering collaboration, ESM3 can continue to revolutionize protein analysis and drive scientific discovery. The next steps for organizations and researchers involve embracing these innovations and contributing to the ongoing evolution of this transformative technology.
13. Building a Community Around ESM3
Creating a thriving community around ESM3 models can drive innovation, foster collaboration, and enhance the accessibility of this transformative technology. This chapter explores strategies for community building, practical examples for engagement, and the benefits of open collaboration in expanding ESM3 applications.
13.1 Importance of Community Building
An active and engaged community is critical for:
- Knowledge Sharing: Facilitating the exchange of best practices and insights.
- Collaboration: Enabling cross-disciplinary projects and research.
- Support System: Helping users troubleshoot and optimize their workflows.
- Innovation: Encouraging contributions that enhance ESM3’s capabilities.
Benefits for Different Stakeholders:
- Researchers: Gain access to datasets, tools, and peer-reviewed methods.
- Developers: Learn optimization techniques and share deployment workflows.
- Enterprises: Collaborate on industry-specific applications and innovations.
13.2 Strategies for Building an ESM3 Community
1. Create Accessible Documentation and Tutorials
Comprehensive documentation lowers the barrier to entry for new users.
Example: Developing Step-by-Step Tutorials
- Beginner tutorials: Installing ESM3, running basic predictions.
- Advanced tutorials: Customizing models, integrating ESM3 into pipelines.
Sample Tutorial Outline:
- Title: "Getting Started with ESM3"
- Section 1: Installing ESM3
- Section 2: Running Your First Prediction
- Section 3: Visualizing Results with Heatmaps
Snippet: Simple ESM3 Prediction Example
pythonCopy codefrom esm3 import ESM3Model
model = ESM3Model()
sequence = "MKTLLILAVVAAALA"
prediction = model.predict(sequence)
print("Prediction Results:")
print(prediction)
2. Organize Workshops and Webinars
Interactive events provide hands-on experience and foster a sense of community.
Key Topics for Workshops:
- Deploying ESM3 in cloud environments.
- Visualizing structural predictions.
- Integrating ESM3 with enterprise applications.
Example Webinar Agenda:
- Introduction to ESM3 (15 minutes)
- Real-world applications (20 minutes)
- Live coding session: Deploying ESM3 with FastAPI (30 minutes)
- Q&A session (15 minutes)
3. Build an Online Knowledge Base
Centralize resources such as FAQs, troubleshooting guides, and use cases.
Example: FAQ Entries
- Q: What are the system requirements for ESM3?
- A: A GPU with CUDA support and at least 16GB of RAM is recommended.
- Q: How do I process large protein datasets?
- A: Use batch processing with optimized data loaders.
4. Foster Open-Source Contributions
Encourage the community to contribute code, plugins, and tools to enhance ESM3’s ecosystem.
Steps for Open-Source Contribution:
- Create a GitHub Repository: Host the ESM3 codebase and include contribution guidelines.
- Encourage Feature Requests: Allow users to suggest features via GitHub Issues.
- Organize Hackathons: Reward innovative solutions and extensions.
Example: Contribution Guidelines
markdownCopy code# Contributing to ESM3
1. Fork the repository.
2. Create a feature branch (`git checkout -b feature/my-feature`).
3. Commit your changes and push to your branch.
4. Open a pull request with a detailed description.
5. Build Collaborative Platforms
Create forums and discussion boards where users can share insights, ask questions, and provide feedback.
Example Platforms:
- Discourse Forums: Host discussions on ESM3 techniques and challenges.
- Slack/Discord Communities: Facilitate real-time collaboration.
Sample Slack Channels:
- #general: Announcements and updates.
- #troubleshooting: Community support for technical issues.
- #deployments: Share deployment strategies and code.
13.3 Community Engagement Tactics
1. Recognize and Reward Contributions
Incentivize contributions through rewards, recognition, and opportunities.
Examples:
- Publish a “Contributor of the Month” blog post highlighting top contributors.
- Offer exclusive access to advanced tutorials or tools for active participants.
2. Host Regular Challenges
Challenges stimulate innovation and provide opportunities for users to demonstrate their skills.
Example Challenge:
- Title: "Optimize ESM3 Predictions for Large Datasets"
- Goal: Develop a scalable pipeline to process a 10,000-sequence dataset.
- Reward: Feature the winning solution in the ESM3 documentation and offer a prize.
3. Provide Continuous Support
Ensure users have access to timely assistance and updates.
Examples:
- Maintain an active GitHub Issues page for bug reporting.
- Publish a monthly newsletter with updates, tutorials, and community highlights.
13.4 Practical Example: Building a Collaborative Knowledge Base
Scenario: An academic consortium wants to centralize ESM3-related resources for researchers across institutions.
Steps to Build the Knowledge Base:
- Platform Selection:
- Use an open-source wiki platform like MediaWiki or Confluence.
- Content Development:
- Add sections for installation guides, use cases, and tutorials.
- Community Contributions:
- Allow registered users to submit articles and updates.
Example Knowledge Base Structure:
- Home: Overview of ESM3.
- Tutorials: Step-by-step guides for beginners and advanced users.
- FAQs: Common questions and answers.
- Resources: Links to datasets, tools, and publications.
- Community: Forums and discussion boards.
Sample Knowledge Base Entry:
markdownCopy code# Visualizing ESM3 Predictions
## Heatmaps for Token Probabilities
Use Matplotlib to create heatmaps:
```python
import matplotlib.pyplot as plt
probabilities = [0.95, 0.89, 0.88, 0.92]
plt.imshow([probabilities], cmap="YlGn", aspect="auto")
plt.colorbar(label="Confidence")
plt.show()
yamlCopy code
---
#### **13.5 Measuring Community Impact**
---
Assessing the success of community-building efforts helps refine strategies and demonstrate value.
---
**Key Metrics to Track:**
- **Engagement:**
- Number of forum posts, GitHub issues, and pull requests.
- **Participation:**
- Attendance at webinars and workshops.
- **Growth:**
- Increase in community members over time.
- **Innovation:**
- Number of new tools, plugins, or workflows developed by the community.
**Example: Tracking Metrics with Google Analytics**
Set up analytics for the knowledge base to track page views, user behavior, and engagement trends.
---
### **Conclusion**
Building a vibrant community around ESM3 can significantly enhance its adoption, accessibility, and innovation. By fostering collaboration, providing resources, and recognizing contributions, the community can drive advancements in protein analysis and structural biology. Future efforts should focus on sustaining engagement and expanding the community to new disciplines and industries.
14. Evaluating the Success of ESM3 Deployments
Deploying ESM3 in production environments or research workflows is a significant achievement, but evaluating the deployment's success is critical for ensuring its effectiveness and identifying areas for improvement. This chapter focuses on key performance metrics, evaluation strategies, and tools to assess the impact of ESM3 deployments.
14.1 Why Evaluate ESM3 Deployments?
Evaluation helps ensure:
- Operational Efficiency: Is the system running optimally in production?
- Model Accuracy: Are predictions aligned with real-world observations?
- Scalability: Can the system handle growing datasets and demands?
- Impact Assessment: Is the deployment achieving its intended outcomes, such as accelerating drug discovery or improving diagnostics?
Example: Evaluation in Drug Discovery
- Objective: Assess whether ESM3’s protein predictions improve drug target identification.
- Evaluation Metric: Reduction in time required to identify potential targets compared to traditional methods.
14.2 Key Performance Metrics for ESM3
1. Prediction Accuracy
- Measure how closely ESM3’s predictions align with experimental or validated data.
Common Metrics:
- Precision and Recall: Evaluate true positive and false positive rates in predictions.
- F1 Score: A harmonic mean of precision and recall.
- RMSE (Root Mean Square Error): For regression-based predictions like structural distances.
Example: Calculating Prediction Accuracy
pythonCopy codefrom sklearn.metrics import precision_score, recall_score, f1_score
true_labels = [1, 0, 1, 1, 0]
predicted_labels = [1, 0, 1, 0, 0]
precision = precision_score(true_labels, predicted_labels)
recall = recall_score(true_labels, predicted_labels)
f1 = f1_score(true_labels, predicted_labels)
print(f"Precision: {precision}, Recall: {recall}, F1 Score: {f1}")
2. Computational Efficiency
- Assess the time and resources required to process predictions.
Key Metrics:
- Inference Time: Average time to generate predictions for a single sequence.
- Resource Utilization: CPU, GPU, and memory usage during deployment.
Example: Logging Inference Times
pythonCopy codeimport time
start_time = time.time()
predictions = esm3_model.predict(sequence)
end_time = time.time()
print(f"Inference Time: {end_time - start_time} seconds")
3. Scalability
- Measure how the system performs under increasing loads.
Key Metrics:
- Throughput: Number of sequences processed per second.
- Latency: Time delay for processing additional requests during high traffic.
Example: Simulating High Loads
pythonCopy codeimport concurrent.futures
sequences = ["SEQ1", "SEQ2", "SEQ3", "SEQ4"]
def process_sequence(sequence):
return esm3_model.predict(sequence)
with concurrent.futures.ThreadPoolExecutor() as executor:
results = list(executor.map(process_sequence, sequences))
print(f"Processed {len(results)} sequences concurrently.")
4. Business or Research Impact
- Evaluate how ESM3 contributes to tangible outcomes, such as cost savings, faster drug discovery, or more accurate diagnoses.
14.3 Evaluation Frameworks
1. Benchmarks
- Compare ESM3 predictions against standard datasets or existing models.
Example: Using Benchmarks for Protein Folding
- Dataset: CASP (Critical Assessment of Protein Structure Prediction).
- Metric: Compare predicted structures to experimental results using TM-score or RMSD.
Python Snippet: Comparing RMSD
pythonCopy codeimport numpy as np
# True and predicted coordinates
true_coords = np.array([[1.0, 1.0, 1.0], [2.0, 2.0, 2.0]])
predicted_coords = np.array([[1.1, 1.1, 1.1], [1.9, 1.9, 1.9]])
rmsd = np.sqrt(np.mean((true_coords - predicted_coords) ** 2))
print(f"RMSD: {rmsd}")
2. A/B Testing
- Use controlled experiments to compare ESM3’s impact against alternative methods.
Example: A/B Testing in Diagnostics
- Group A: Use ESM3 for protein mutation analysis.
- Group B: Use traditional analysis methods.
- Compare accuracy, time-to-result, and diagnostic effectiveness.
3. Real-Time Monitoring
- Monitor deployments in production to evaluate ongoing performance.
Key Tools:
- Prometheus: Monitor metrics like latency and resource usage.
- Grafana: Visualize performance data through dashboards.
Example: Monitoring API Latency
yamlCopy code# Prometheus configuration snippet
- job_name: "esm3_api"
static_configs:
- targets: ["localhost:8000"]
4. Feedback Loops
- Collect user feedback to identify issues and areas for improvement.
Example: User Feedback Form
- Create a simple feedback mechanism in your application:
pythonCopy codeimport json
feedback = {
"user": "researcher1",
"use_case": "drug discovery",
"comments": "The predictions are accurate but could be faster."
}
with open("feedback.json", "w") as f:
json.dump(feedback, f)
14.4 Tools for Evaluation
1. Model Evaluation Libraries
- Scikit-learn: For evaluating accuracy metrics.
- BioPython: For analyzing biological data outputs.
2. Profiling Tools
- cProfile: Python tool to profile computational bottlenecks.
- NVIDIA Nsight: Analyze GPU performance.
3. Cloud-Based Analytics
- AWS CloudWatch: Monitor deployments on AWS.
- Azure Monitor: Evaluate performance on Microsoft Azure.
14.5 Practical Example: End-to-End Evaluation
Scenario: A pharmaceutical company deployed ESM3 to predict drug-binding sites for proteins. The deployment’s success is evaluated across multiple metrics.
Steps:
- Set Up Benchmarks:
- Use a validated dataset of protein-ligand interactions.
- Measure Computational Efficiency:
- Log inference times and GPU utilization.
- Monitor Scalability:
- Simulate batch processing with 1,000 sequences.
- Assess Impact:
- Calculate the time saved compared to traditional methods.
Code Implementation:
pythonCopy code# Step 1: Benchmark Predictions
benchmark_data = load_benchmark_data()
for protein in benchmark_data:
prediction = esm3_model.predict(protein["sequence"])
compare_results(protein["expected_binding_site"], prediction["binding_site"])
# Step 2: Log Efficiency
import time
start_time = time.time()
esm3_model.predict("MKTLLILAVVAAALA")
print(f"Inference Time: {time.time() - start_time} seconds")
# Step 3: Simulate Scalability
from concurrent.futures import ThreadPoolExecutor
sequences = ["SEQ" + str(i) for i in range(1000)]
def batch_process(sequence):
return esm3_model.predict(sequence)
with ThreadPoolExecutor() as executor:
results = list(executor.map(batch_process, sequences))
# Step 4: Assess Impact
print("Time saved: 40% compared to traditional methods.")
Evaluating ESM3 deployments ensures that they deliver on their promises of accuracy, efficiency, and scalability. By leveraging metrics, evaluation frameworks, and tools, you can continuously refine your deployment and maximize its impact. This comprehensive evaluation approach provides actionable insights, paving the way for iterative improvements and long-term success.
15. Conclusion and Recommendations for Future ESM3 Deployments
Deploying ESM3 models in production environments marks a significant milestone in leveraging advanced protein analysis tools for research and industry. This concluding chapter synthesizes the key insights from earlier discussions, identifies best practices, and outlines future recommendations for maximizing the potential of ESM3.
15.1 Summary of Key Insights
- Model Preparation and Customization
- Ensure the ESM3 model is fine-tuned for specific datasets to improve prediction accuracy.
- Leverage tools like embeddings clustering and sequence-level analysis to extract meaningful insights.
- Deployment Strategies
- Utilize scalable frameworks like Kubernetes for managing workloads.
- Optimize inference pipelines using batching and hardware acceleration.
- Evaluation and Monitoring
- Measure success through metrics such as prediction accuracy, inference speed, and impact on research outcomes.
- Use monitoring tools like Prometheus and Grafana to track performance in real time.
- Community and Collaboration
- Build knowledge-sharing platforms to encourage contributions and collaborative problem-solving.
- Engage in interdisciplinary partnerships to expand ESM3’s applications.
- Future Trends
- Prepare for innovations such as multimodal integration, federated learning, and edge computing.
15.2 Best Practices for ESM3 Deployments
1. Understand Your Objectives
- Clearly define the purpose of the deployment (e.g., drug discovery, protein engineering).
- Align goals with measurable success criteria, such as time savings or improved accuracy.
Example: Goal Setting for Drug Discovery
- Objective: Identify 5 novel drug targets within 3 months.
- Metrics: Precision and recall of binding site predictions, time saved compared to manual methods.
2. Optimize Model Performance
- Regularly update ESM3 to leverage the latest advancements.
- Use dimensionality reduction techniques to handle high-dimensional embeddings.
Example: Reducing Embedding Dimensions for Faster Processing
pythonCopy codefrom sklearn.decomposition import PCA
import numpy as np
# Example embeddings
embeddings = np.random.rand(100, 768)
# Reduce dimensions
pca = PCA(n_components=50)
reduced_embeddings = pca.fit_transform(embeddings)
print(f"Reduced embeddings shape: {reduced_embeddings.shape}")
3. Build Resilient Pipelines
- Incorporate error handling for issues like incomplete datasets or failed predictions.
- Use retries and fallbacks to maintain pipeline robustness.
Example: Adding Fallbacks in Prediction Pipelines
pythonCopy codedef predict_with_fallback(sequence):
try:
return esm3_model.predict(sequence)
except Exception as e:
print(f"Prediction failed for {sequence}: {e}")
return {"error": "Failed prediction", "sequence": sequence}
prediction = predict_with_fallback("MKTLLILAVVAAALA")
4. Engage with the Community
- Regularly contribute to forums and GitHub repositories to share findings and gather insights.
- Participate in hackathons or workshops to stay updated on best practices.
15.3 Recommendations for Future Deployments
1. Prioritize Accessibility
- Simplify deployment processes to make ESM3 accessible to non-technical users.
- Develop user-friendly tools and GUIs for running predictions and visualizations.
Example: Building a GUI for ESM3
pythonCopy codeimport tkinter as tk
from tkinter import filedialog
from esm3 import ESM3Model
def predict_sequence():
sequence = sequence_entry.get()
prediction = esm3_model.predict(sequence)
result_label.config(text=str(prediction))
# GUI setup
root = tk.Tk()
root.title("ESM3 Prediction Tool")
tk.Label(root, text="Enter Protein Sequence:").pack()
sequence_entry = tk.Entry(root, width=50)
sequence_entry.pack()
predict_button = tk.Button(root, text="Predict", command=predict_sequence)
predict_button.pack()
result_label = tk.Label(root, text="")
result_label.pack()
root.mainloop()
2. Expand Interdisciplinary Applications
- Explore new domains such as environmental science, materials engineering, or personalized medicine.
- Collaborate with experts from different fields to identify novel use cases.
3. Integrate Advanced Technologies
- Incorporate AI techniques like attention visualization to improve interpretability.
- Use federated learning to train models on sensitive datasets while preserving privacy.
Example: Federated Learning with ESM3
pythonCopy codefrom federated_learning import FederatedModel
# Define a federated model
federated_esm3 = FederatedModel(base_model=esm3_model, clients=["lab1", "lab2"])
federated_esm3.train()
4. Focus on Sustainability
- Optimize resource usage to reduce energy consumption.
- Design workflows that are scalable and efficient for large datasets.
Example: Resource Optimization with Batch Processing
pythonCopy codedef batch_predict(sequences, batch_size):
for i in range(0, len(sequences), batch_size):
batch = sequences[i:i + batch_size]
yield esm3_model.predict_batch(batch)
sequences = ["SEQ1", "SEQ2", "SEQ3", "SEQ4"]
for predictions in batch_predict(sequences, batch_size=2):
print(predictions)
15.4 Practical Example: End-to-End Deployment
Scenario:
A biotechnology company aims to deploy ESM3 for high-throughput analysis of protein sequences to identify enzymes for industrial applications.
Steps:
- Preparation:
- Fine-tune ESM3 on industrial enzyme datasets.
- Validate predictions using benchmark datasets.
- Deployment:
- Set up an inference pipeline on AWS using Lambda and Docker.
- Use batch processing to handle large datasets.
- Evaluation:
- Monitor accuracy, throughput, and resource utilization.
- Adjust parameters based on feedback from researchers.
- Knowledge Sharing:
- Publish a case study highlighting results and lessons learned.
Implementation:
pythonCopy code# Fine-tune ESM3
fine_tuned_model = esm3_model.fine_tune("enzyme_dataset")
# Deploy with Docker
import docker
client = docker.from_env()
container = client.containers.run("esm3_model_image", ports={"8080/tcp": 8080}, detach=True)
# Batch processing pipeline
sequences = ["SEQ1", "SEQ2", "SEQ3"]
batch_size = 2
def process_in_batches(sequences, batch_size):
for i in range(0, len(sequences), batch_size):
batch = sequences[i:i + batch_size]
yield esm3_model.predict_batch(batch)
for predictions in process_in_batches(sequences, batch_size):
print(predictions)
15.5 Final Thoughts
ESM3 represents a groundbreaking advancement in protein analysis, offering immense potential for research and industry. To fully leverage its capabilities:
- Continuously innovate and optimize deployments.
- Engage with the global community to share knowledge and drive improvements.
- Expand applications into new domains to address pressing scientific and industrial challenges.
By following best practices and embracing future innovations, organizations can maximize the impact of ESM3, contributing to significant breakthroughs in computational biology and beyond.
Visited 1 times, 1 visit(s) today
Leave a Reply