Chapter 1: What is ESM3?
Introduction: The Cutting-Edge Frontier of AI
In recent years, artificial intelligence has achieved remarkable milestones, transforming industries and advancing scientific discovery. Among these innovations is ESM3, a revolutionary model designed to tackle some of the most complex problems in bioinformatics, natural language processing, and beyond. Developed with precision and insight, ESM3 bridges the gap between high-dimensional data and actionable insights, enabling breakthroughs in areas like protein folding and genomic research.
This chapter introduces ESM3, explaining its core architecture and practical implications. Designed to demystify its complexities, this chapter provides a foundation for understanding the neural network layers of ESM3 and sets the stage for exploring its detailed mechanisms in subsequent chapters.
What is ESM3?
At its core, ESM3 is a neural network model based on transformer architecture, optimized for solving highly specialized tasks. Initially developed to process sequences, ESM3 excels in tasks such as protein structure prediction, where precise relationships within sequence data are critical. By leveraging self-attention mechanisms and deep contextual embeddings, ESM3 outperforms traditional models in both accuracy and scalability.
The Evolution of AI Models
To understand ESM3, it’s essential to consider its place in the evolution of AI models:
- Rule-Based Systems: Early AI systems relied on handcrafted rules. While effective for static tasks, these systems lacked adaptability.
- Machine Learning Models: Models such as support vector machines (SVMs) and decision trees automated pattern recognition but required feature engineering.
- Deep Learning Models: Neural networks introduced feature learning, with architectures like convolutional neural networks (CNNs) excelling in image processing and recurrent neural networks (RNNs) dominating sequential data tasks.
- Transformers: Introduced by Vaswani et al. in 2017, transformers revolutionized AI by introducing self-attention mechanisms that effectively handle sequence data without recurrence.
ESM3 is a direct descendant of this evolutionary trajectory, designed to address the nuanced challenges of bioinformatics and sequence processing.
Applications of ESM3
The practical utility of ESM3 extends across multiple disciplines:
- Protein Folding: Predicting protein structures is critical for understanding biological functions and developing drugs. ESM3 provides high-resolution predictions, streamlining research pipelines.
- Genomics: ESM3 helps identify patterns in genetic sequences, contributing to advancements in personalized medicine and evolutionary studies.
- Natural Language Understanding: While primarily used in scientific domains, ESM3’s architecture can be adapted for NLP tasks, enabling cross-disciplinary applications.
The Role of Open-Source Accessibility
One of the most empowering aspects of ESM3 is its open-source nature. By making cutting-edge technology accessible to researchers worldwide, ESM3 democratizes innovation. Anyone with a basic understanding of AI and programming can experiment with the model, adapt it for their needs, and contribute to its ongoing development.
Understanding the Foundations: How ESM3 Works
At a high level, ESM3 processes input data through several stages, each designed to extract, refine, and contextualize information. These stages correspond to the neural network layers that we’ll explore in depth in later chapters. For now, let’s outline the general workflow:
- Input Preprocessing: ESM3 tokenizes and encodes input sequences into a numerical format suitable for processing.
- Embedding Layer: Converts tokens into high-dimensional vector representations, capturing semantic and structural information.
- Attention Mechanism: Applies self-attention to determine relationships between sequence elements.
- Intermediate Layers: Refines these relationships through multiple transformer blocks, deepening the contextual understanding.
- Output Layer: Produces predictions or embeddings tailored to specific tasks.
Key Features of ESM3
- Transformer-Based Architecture: Unlike older sequence models like RNNs, ESM3 uses transformers for parallel processing, significantly increasing efficiency.
- Multi-Head Attention: ESM3’s attention mechanism allows it to focus on multiple relationships simultaneously, offering a nuanced understanding of input data.
- Scalability: ESM3 can handle large-scale datasets efficiently, making it ideal for complex tasks like analyzing entire genomes.
- Customizability: Researchers can fine-tune ESM3 for domain-specific problems, ensuring optimal performance.
Example: Protein Folding with ESM3
Consider the task of predicting a protein’s 3D structure from its amino acid sequence. ESM3’s workflow might look like this:
- Input: A sequence of amino acids is tokenized into a numerical format.
- Embedding Layer: Each token is mapped to a high-dimensional vector representing its chemical properties.
- Attention Mechanism: Self-attention identifies interactions between amino acids critical for folding.
- Output: The model predicts the spatial arrangement of the protein, which can be visualized using molecular modeling tools.
This example demonstrates ESM3’s ability to handle complex, domain-specific challenges.
Mathematical Foundations of ESM3
The heart of ESM3 lies in its attention mechanism, which can be described mathematically:
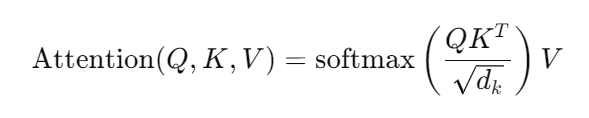
Attention(Q,K,V)=softmax(QKTdk)V\text{Attention}(Q, K, V) = \text{softmax}\left(\frac{QK^T}{\sqrt{d_k}}\right)VAttention(Q,K,V)=softmax(dkQKT)V
Where:
- QQQ: Query matrix.
- KKK: Key matrix.
- VVV: Value matrix.
- dkd_kdk: Dimensionality of the keys.
This formula ensures that the model focuses on relevant parts of the input sequence, balancing efficiency and accuracy.
Code Snippet: Loading ESM3
Below is an example of loading and using ESM3 for sequence analysis:
pythonCopy codefrom esm import pretrained, Alphabet
# Load the ESM3 model
model, alphabet = pretrained.esm3_model()
batch_converter = alphabet.get_batch_converter()
# Example sequence
sequences = [("Protein1", "MTEYKLVVVGAGGVGKSALTIQLIQNHFVDEYDPTIEDSYRKQVVIDGETCLLDILDTAG"),
("Protein2", "GDVAKGEPVQLVCDNGSGLVQINKLKCLIEKFTKDYGVKTKQIKLHGLENVRDYLIP")]
# Preprocess sequences
batch_labels, batch_strs, batch_tokens = batch_converter(sequences)
# Predict outputs
with torch.no_grad():
results = model(batch_tokens)
This snippet shows how easily researchers can integrate ESM3 into their workflows.
Looking Ahead
With a foundational understanding of ESM3’s purpose, architecture, and applications, the next chapters will delve deeper into its neural network layers. From embedding layers to attention mechanisms, each component will be explored in detail, complete with examples and use cases. By the end of this book, you’ll have the knowledge and tools to harness the full potential of ESM3 for your research or projects.
Chapter 2: The Basics of Neural Networks
Introduction: Setting the Stage
Neural networks form the backbone of modern artificial intelligence, driving advancements in fields as diverse as image recognition, natural language processing, and bioinformatics. Before diving deep into the neural network architecture of ESM3, it is essential to grasp the fundamentals of neural networks. This chapter provides a detailed yet accessible exploration of neural networks, focusing on the foundational principles and their application to sequence-based models like ESM3.
By the end of this chapter, readers will have a solid understanding of how neural networks process data, the importance of layers in extracting meaningful patterns, and why models like ESM3 represent a significant evolution in AI.
What Are Neural Networks?
At their core, neural networks are computational models inspired by the human brain. They consist of interconnected nodes (neurons) organized into layers, mimicking the way biological neurons transmit information. Each node processes input data, applies a transformation (usually a weighted sum followed by a non-linear function), and passes the result to the next layer.
Key Components:
- Input Layer: The first layer receives raw data, transforming it into a format the network can process.
- Hidden Layers: Intermediate layers that apply transformations to capture complex patterns in the data.
- Output Layer: The final layer produces the network’s prediction or decision.
How Neural Networks Process Data
A neural network processes data through three main stages:
- Forward Propagation:
- Data flows through the network from the input layer to the output layer.
- Each neuron applies a transformation: z=∑i=1nwixi+bz = \sum_{i=1}^{n} w_i x_i + bz=i=1∑nwixi+b Here:
- zzz: The neuron’s output.
- wiw_iwi: Weight for input xix_ixi.
- bbb: Bias term.
- The result passes through an activation function, such as:
- Sigmoid: σ(z)=11+e−z\sigma(z) = \frac{1}{1 + e^{-z}}σ(z)=1+e−z1
- ReLU: f(z)=max(0,z)f(z) = \max(0, z)f(z)=max(0,z)
- Loss Calculation:
- The network compares its output to the actual target using a loss function, e.g., mean squared error (MSE) or cross-entropy.
- Backward Propagation:
- The network adjusts its weights and biases to minimize the loss, using algorithms like gradient descent: wi←wi−η∂L∂wiw_i \leftarrow w_i – \eta \frac{\partial L}{\partial w_i}wi←wi−η∂wi∂L Where η\etaη is the learning rate, and LLL is the loss.
Why Layers Matter
The depth and structure of layers determine the network’s ability to extract meaningful patterns. In sequence-based models like ESM3:
- Shallow Networks: Capture simple, linear relationships.
- Deep Networks: Capture hierarchical, non-linear patterns, making them well-suited for tasks like protein structure prediction.
From Traditional Models to Transformers
Traditional neural network architectures like recurrent neural networks (RNNs) and convolutional neural networks (CNNs) laid the groundwork for modern AI. However, they struggled with certain challenges, such as vanishing gradients and processing long sequences. Transformers, which power ESM3, addressed these limitations.
Key Innovations of Transformers:
- Self-Attention Mechanism:
- Allows the model to focus on relationships between elements in a sequence.
- Formula: Attention(Q,K,V)=softmax(QKTdk)V\text{Attention}(Q, K, V) = \text{softmax}\left(\frac{QK^T}{\sqrt{d_k}}\right)VAttention(Q,K,V)=softmax(dkQKT)V
- Here, QQQ, KKK, and VVV represent the query, key, and value matrices.
- Parallel Processing:
- Unlike RNNs, transformers process sequences in parallel, significantly improving computational efficiency.
- Scalability:
- With multi-head attention and stacked layers, transformers like ESM3 scale to handle complex tasks efficiently.
Example: Neural Networks in Protein Sequence Analysis
To illustrate how neural networks process sequence data, let’s consider the task of predicting the functional regions of a protein:
- Input:
- Amino acid sequence:
MTEYKLVVVGAGGVGKSALTIQLIQNHFVDE
- Amino acid sequence:
- Embedding:
- The input sequence is tokenized, and each token is mapped to a high-dimensional vector.
- Forward Propagation:
- The embedding vectors pass through multiple layers of the network, where each layer refines the representation by capturing interdependencies among amino acids.
- Output:
- The network predicts regions critical for the protein’s function, such as binding sites.
Building Blocks of ESM3’s Neural Network
While the general principles of neural networks apply to all architectures, ESM3 introduces unique features tailored to sequence analysis:
- Positional Encoding:
- Captures the order of sequence elements, ensuring the model understands sequence-dependent relationships.
- Formula: PE(pos,2i)=sin(pos100002i/d)PE(pos, 2i) = \sin\left(\frac{pos}{10000^{2i/d}}\right)PE(pos,2i)=sin(100002i/dpos) PE(pos,2i+1)=cos(pos100002i/d)PE(pos, 2i+1) = \cos\left(\frac{pos}{10000^{2i/d}}\right)PE(pos,2i+1)=cos(100002i/dpos)
- Transformer Blocks:
- Stacked layers combining self-attention and feedforward components, allowing the model to extract hierarchical patterns.
- Output Representations:
- Produces embeddings or predictions tailored to tasks like protein folding or genomic analysis.
Code Example: Simple Neural Network for Sequence Data
Below is an example of implementing a basic neural network for sequence classification using PyTorch:<pre><code> import torch import torch.nn as nn import torch.optim as optim # Define the neural network class SequenceClassifier(nn.Module): def __init__(self, input_size, hidden_size, output_size): super(SequenceClassifier, self).__init__() self.hidden_layer = nn.Linear(input_size, hidden_size) self.output_layer = nn.Linear(hidden_size, output_size) self.activation = nn.ReLU() def forward(self, x): hidden = self.activation(self.hidden_layer(x)) output = self.output_layer(hidden) return output # Initialize the network model = SequenceClassifier(input_size=100, hidden_size=50, output_size=2) criterion = nn.CrossEntropyLoss() optimizer = optim.Adam(model.parameters(), lr=0.001) # Example data input_data = torch.randn(32, 100) # Batch of 32 sequences, each of length 100 labels = torch.randint(0, 2, (32,)) # Binary classification labels # Training step optimizer.zero_grad() outputs = model(input_data) loss = criterion(outputs, labels) loss.backward() optimizer.step() </code></pre>
Looking Ahead
This chapter established a foundational understanding of neural networks, emphasizing their role in processing sequence data. The next chapter will dive deeper into the first layer of ESM3: the Input Layer. Readers will learn how ESM3 translates raw data into a format suitable for neural network processing, setting the stage for its advanced capabilities.
Chapter 3: ESM3 at a Glance – Architecture Overview
Introduction: The Blueprint of Innovation
ESM3 stands as a revolutionary leap in the application of transformer-based neural networks, purpose-built for handling complex sequence data. Its architecture reflects years of research and innovation, optimizing computational efficiency while delivering groundbreaking results in tasks like protein folding and genomic analysis. This chapter delves into the architectural design of ESM3, exploring its modular components and how they interconnect to process and interpret data.
We’ll dissect each core component, highlighting the significance of design choices that set ESM3 apart from conventional neural networks. By the end of this chapter, you’ll have a clear understanding of ESM3’s architecture, paving the way for deeper exploration of its layers.
Overview of ESM3’s Architecture
At its heart, ESM3 is a transformer-based architecture specifically optimized for sequence modeling. It builds upon the success of transformers like BERT and GPT, adapting and enhancing their designs for biological and scientific applications.
Key Features of ESM3:
- Sequence-Aware Design: Tailored for tasks requiring deep contextual understanding of sequences, such as amino acids or genomic data.
- Scalability: Handles large datasets efficiently, enabling the analysis of complex biological systems.
- Modularity: Each component is designed to perform a specific function, ensuring flexibility and adaptability for various use cases.
High-Level Workflow:
- Input Preprocessing
- Embedding Layer
- Multi-Head Self-Attention
- Feedforward Layers
- Output Representations
Component Breakdown
1. Input Preprocessing
Input preprocessing ensures that raw data is transformed into a format suitable for neural network processing. For sequence data, preprocessing includes:
- Tokenization: Breaking down sequences (e.g., amino acids) into individual tokens.
- Numerical Encoding: Mapping tokens to numerical values that the model can interpret.
Example: For the protein sequence MTEYKLVVVGAGGVGKSALTIQLIQNHFVDE, preprocessing might generate a tokenized representation like: [M, T, E, Y, K, L, V, V, V, G, A, G, G, V, G, K, S, A, L, T, I, Q, L, I, Q, N, H, F, V, D, E]
These tokens are then numerically encoded using a vocabulary defined during training.
2. Embedding Layer
The embedding layer maps each token into a high-dimensional space, capturing both semantic and structural information. This step is crucial for sequence tasks where context and order matter.Embedding(xi)=Token Vector+Positional Encoding\text{Embedding}(x_i) = \text{Token Vector} + \text{Positional Encoding}Embedding(xi)=Token Vector+Positional Encoding
- Token Vector: Represents the inherent properties of each token.
- Positional Encoding: Adds information about the token’s position in the sequence.
Example Visualization:
- Input:
MTE - Embedding Vector: A 768-dimensional vector for each token, e.g.,
M: [0.23, -0.11, ...], T: [0.12, 0.34, ...], E: [-0.45, 0.67, ...]
3. Multi-Head Self-Attention
The self-attention mechanism enables ESM3 to focus on relationships between tokens within a sequence, identifying dependencies that may span long distances.
Mathematical Representation:Attention(Q,K,V)=softmax(QKTdk)V\text{Attention}(Q, K, V) = \text{softmax}\left(\frac{QK^T}{\sqrt{d_k}}\right)VAttention(Q,K,V)=softmax(dkQKT)V
- QQQ: Query matrix derived from the input.
- KKK: Key matrix representing contextual information.
- VVV: Value matrix for token significance.
Each attention head operates independently, and their outputs are concatenated and linearly transformed to produce the final result.
4. Feedforward Layers
Feedforward layers refine the representation further by applying non-linear transformations. These layers consist of:
- A linear transformation with ReLU activation: ReLU(x)=max(0,x)\text{ReLU}(x) = \max(0, x)ReLU(x)=max(0,x)
- Another linear transformation to project back into the embedding space.
This mechanism enhances the model’s ability to capture complex patterns.
5. Output Representations
The final layer produces task-specific outputs:
- For classification: Outputs probabilities over predefined categories.
- For embedding tasks: Generates vector representations suitable for downstream analysis.
Example: In a protein folding task, the output might represent the 3D coordinates of the protein structure.
How ESM3 Improves Over Traditional Models
- Efficiency in Handling Long Sequences: ESM3 processes sequences in parallel using self-attention, unlike RNNs, which require sequential computation.
- Rich Contextual Understanding: Multi-head attention enables the model to simultaneously focus on multiple relationships, crucial for tasks like protein interaction mapping.
- Scalability: ESM3’s modular design ensures that it can be fine-tuned or extended to handle large-scale datasets.
Practical Use Case: Protein Structure Prediction
Predicting the 3D structure of a protein involves mapping a sequence of amino acids to its spatial configuration. ESM3 achieves this by:
- Tokenizing the Input Sequence: Breaking down the sequence into amino acids.
- Embedding: Creating high-dimensional vectors representing each amino acid.
- Attention Mechanisms: Identifying dependencies between amino acids critical for folding.
- Output: Predicting spatial coordinates or interaction probabilities.
Code Example: Visualizing Attention Weights
Below is an example of how to extract and visualize attention weights using ESM3:<pre><code> from esm import pretrained, Alphabet import torch import matplotlib.pyplot as plt # Load ESM3 model model, alphabet = pretrained.esm3_model() batch_converter = alphabet.get_batch_converter() # Example sequence sequences = [(“Protein1”, “MTEYKLVVVGAGGVGKSALTIQLIQNHFVDE”)] # Preprocess sequence batch_labels, batch_strs, batch_tokens = batch_converter(sequences) # Extract attention weights with torch.no_grad(): outputs = model(batch_tokens, return_attentions=True) attention_weights = outputs[‘attentions’] # Visualize attention weights for first layer and head plt.imshow(attention_weights[0][0].detach().numpy(), cmap=’viridis’) plt.title(“Attention Weights”) plt.xlabel(“Sequence Position”) plt.ylabel(“Sequence Position”) plt.colorbar() plt.show() </code></pre>
Looking Ahead
This chapter outlined ESM3’s architecture, highlighting the design choices that make it a powerful tool for sequence modeling. The next chapter will explore the first layer in detail: the Input Layer. Readers will learn how ESM3 tokenizes and encodes raw sequences, preparing them for deeper analysis.
Chapter 4: The Input Layer – Translating Data for ESM3
Introduction: Where Data Meets Neural Networks
Every journey in a neural network begins at the input layer, where raw data is transformed into a format that the model can process. In the case of ESM3, this step is especially critical. The input layer bridges the gap between the biological or textual representation of sequence data and the numerical abstractions required by machine learning models. In this chapter, we’ll explore the intricacies of ESM3’s input layer, including tokenization, encoding, and preprocessing.
Through examples, code snippets, and detailed explanations, this chapter provides a clear understanding of how ESM3 prepares data for its downstream neural network layers.
The Role of the Input Layer
The input layer’s primary responsibility is to preprocess and encode raw data into a structured form that the neural network can interpret. In ESM3, this involves:
- Tokenization: Breaking down sequences into smaller components (tokens) for processing.
- Numerical Encoding: Mapping tokens into numerical indices.
- Batch Preparation: Structuring the input data to optimize for computational efficiency.
Why It Matters
Unlike standard text models, ESM3 often deals with highly complex and structured data, such as protein sequences. Proper preprocessing ensures that the model captures the relationships and dependencies within this data without losing critical context.
Tokenization in ESM3
Tokenization is the process of splitting a sequence into discrete units, or tokens, that the model can interpret. For biological sequences, tokens may represent:
- Amino Acids: Individual letters in a protein sequence.
- Nucleotides: Bases in a DNA or RNA sequence.
Example: Protein Sequence
Input Sequence:MTEYKLVVVGAGGVGKSALTIQLIQNHFVDE
Tokenized Output:[M, T, E, Y, K, L, V, V, V, G, A, G, G, V, G, K, S, A, L, T, I, Q, L, I, Q, N, H, F, V, D, E]
Numerical Encoding
Once tokenized, sequences are converted into numerical indices using a predefined vocabulary. This step maps each token to an integer, enabling the model to process the data efficiently.
Vocabulary Example:
| Token | Index |
|---|---|
| M | 1 |
| T | 2 |
| E | 3 |
| … | … |
For the tokenized sequence [M, T, E, Y], the numerical encoding would be [1, 2, 3, 25].
Positional Encoding
Sequence models like ESM3 must account for the order of tokens, as their position affects the model’s interpretation. To achieve this, ESM3 employs positional encodings—vectors added to each token embedding to capture positional information.
Formula for Positional Encoding:
PE(pos,2i)=sin(pos100002i/d)PE(pos, 2i) = \sin\left(\frac{pos}{10000^{2i/d}}\right)PE(pos,2i)=sin(100002i/dpos) PE(pos,2i+1)=cos(pos100002i/d)PE(pos, 2i+1) = \cos\left(\frac{pos}{10000^{2i/d}}\right)PE(pos,2i+1)=cos(100002i/dpos)
Here:
- pospospos: Position of the token in the sequence.
- iii: Dimension index.
- ddd: Total embedding dimension.
Visual Representation:
For a sequence of length 5, positional encodings may look like this:
| Position | Dimension 1 (sin) | Dimension 2 (cos) | … |
|---|---|---|---|
| 1 | 0.841 | 0.540 | … |
| 2 | 0.909 | 0.416 | … |
| … | … | … | … |
Batch Preparation
To maximize computational efficiency, ESM3 processes data in batches. Batch preparation involves:
- Padding: Adding dummy tokens to sequences of varying lengths to ensure uniformity.
- Masking: Identifying padded tokens so they are ignored during computation.
Example:
| Sequence | Length | Padded Sequence |
|---|---|---|
MTEYK | 5 | MTEYK0000 |
VVVG | 4 | VVVG00000 |
Mask: [1, 1, 1, 1, 1, 0, 0, 0] (1 for real tokens, 0 for padding).
Real-World Application: Protein Structure Prediction
Let’s consider a practical example: predicting the structure of a protein from its sequence.
- Input Sequence:
MTEYKLVVVGAGGVGKSALTIQLIQNHFVDE - Preprocessing:
- Tokenization:
[M, T, E, Y, K, ...] - Encoding:
[1, 2, 3, 25, 11, ...] - Batching: Adding padding and creating masks.
- Tokenization:
- Feeding to ESM3: The processed data is passed to the embedding layer, where token vectors are combined with positional encodings for further processing.
Code Example: Input Preprocessing in ESM3
Below is a Python example demonstrating how to preprocess data for ESM3:<pre><code> from esm import pretrained, Alphabet # Load ESM3 model and utilities model, alphabet = pretrained.esm3_model() batch_converter = alphabet.get_batch_converter() # Input sequences sequences = [(“Protein1”, “MTEYKLVVVGAGGVGKSALTIQLIQNHFVDE”), (“Protein2”, “VVVGAGGVG”)] # Preprocess sequences batch_labels, batch_strs, batch_tokens = batch_converter(sequences) # Output tokens and masks print(“Tokenized Batch:”, batch_tokens) </code></pre>
How Input Preprocessing Drives Performance
Effective preprocessing ensures that:
- Accuracy Improves: Proper tokenization and encoding help the model capture nuanced relationships.
- Efficiency Increases: Uniform batch sizes optimize GPU utilization during training and inference.
- Flexibility Enhances: A modular preprocessing pipeline allows researchers to adapt ESM3 for diverse applications, from protein folding to genomic analysis.
Common Challenges and Solutions
- Challenge: Handling long sequences that exceed the model’s maximum input size.
Solution: Use sliding windows or chunk sequences into manageable segments. - Challenge: Preserving rare tokens in biological data.
Solution: Extend the vocabulary or use embeddings that support rare token handling.
Looking Ahead
This chapter detailed how the input layer transforms raw sequence data into a format compatible with ESM3’s neural network. Next, we’ll explore the embedding layer, where numerical encodings are transformed into rich, high-dimensional vectors that capture semantic and structural relationships.
Chapter 5: The Embedding Layer – Representing the Input
Introduction: Transforming Data into Meaning
In the journey through ESM3’s neural network layers, the embedding layer serves as the first true transformation stage. Here, tokenized and encoded inputs are converted into high-dimensional vector representations that encapsulate their semantic and structural meaning. For sequence-based models like ESM3, this layer is critical to understanding the relationships and dependencies between elements, whether they are amino acids, nucleotides, or other sequence data.
This chapter delves into the mechanics of the embedding layer, illustrating how it bridges raw input and the model’s deeper layers. With examples, code snippets, and real-world applications, we’ll explore how ESM3’s embeddings unlock the potential of sequence data.
What is an Embedding Layer?
An embedding layer is a specialized neural network layer that maps discrete inputs (e.g., tokens) to continuous, dense vector representations. Each token is associated with a vector in a high-dimensional space, and tokens with similar meanings or relationships are mapped closer together in this space.
Key Characteristics:
- Dimensionality: Typically, embeddings have hundreds to thousands of dimensions.
- Contextual Representation: Embeddings capture both the inherent properties of tokens and their relationships within a sequence.
- Compactness: Dense vectors represent complex information more efficiently than sparse, one-hot encodings.
How the Embedding Layer Works
The embedding layer processes input data in two steps:
- Mapping Tokens to Vectors: Each token is assigned an embedding vector based on a pre-trained or trainable matrix.
- Adding Positional Encodings: The vectors are augmented with positional information to retain sequence order.
Mathematics of Embeddings
For a sequence of tokens [t1,t2,…,tn][t_1, t_2, \ldots, t_n][t1,t2,…,tn], the embedding layer produces a matrix EEE of shape n×dn \times dn×d, where nnn is the sequence length and ddd is the embedding dimension.E=[e1,e2,…,en]E = [e_1, e_2, \ldots, e_n]E=[e1,e2,…,en]
Where eie_iei is the embedding vector for token tit_iti, retrieved from an embedding matrix WWW:ei=W[ti]e_i = W[t_i]ei=W[ti]
Positional Encoding Addition:
To incorporate sequence information, positional encodings PEPEPE are added to the embeddings:E′=E+PEE’ = E + PEE′=E+PE
Visualization of Embeddings
Imagine embedding vectors in a 3D space (simplified from hundreds of dimensions). Tokens like M, T, and E in a protein sequence might cluster based on shared properties (e.g., hydrophobicity).
| Token | Embedding Vector |
|---|---|
| M | [0.25, 0.13, -0.67, …] |
| T | [-0.45, 0.88, 0.33, …] |
| E | [0.12, -0.50, 0.44, …] |
These vectors, when visualized, reveal patterns and groupings reflecting biological or contextual similarities.
Importance of the Embedding Layer in ESM3
The embedding layer in ESM3 is tailored for sequence data, ensuring that:
- Structural Information is Preserved: Relationships between amino acids are encoded effectively.
- Scalability is Achieved: High-dimensional embeddings handle large datasets without compromising performance.
- Transfer Learning is Enabled: Pre-trained embeddings provide a foundation for fine-tuning on specific tasks.
Practical Use Case: Protein Sequence Analysis
Consider the task of analyzing protein sequences to predict functional regions.
- Input Sequence:
MTEYKLVVVGAGGVGKSALTIQLIQNHFVDE - Tokenization and Encoding:
- Tokens:
[M, T, E, Y, K, L, V, ...] - Encodings:
[1, 2, 3, 25, 11, 12, 22, ...]
- Tokens:
- Embedding Layer Output: The embedding layer maps each token to a high-dimensional vector:TokenEmbedding Vector (Simplified)M[0.25, 0.13, -0.67, …]T[-0.45, 0.88, 0.33, …]E[0.12, -0.50, 0.44, …]
- Positional Encoding: Positional information is added, ensuring the sequence’s structure is preserved.
Code Example: Working with Embeddings in ESM3
Below is a Python example demonstrating how to extract and visualize embeddings in ESM3:<pre><code> from esm import pretrained, Alphabet import torch # Load ESM3 model and utilities model, alphabet = pretrained.esm3_model() batch_converter = alphabet.get_batch_converter() # Example sequences sequences = [(“Protein1”, “MTEYKLVVVGAGGVGKSALTIQLIQNHFVDE”)] # Preprocess sequence batch_labels, batch_strs, batch_tokens = batch_converter(sequences) # Extract embeddings with torch.no_grad(): results = model(batch_tokens, repr_layers=[33]) # Get embeddings from the last layer embeddings = results[“representations”][33] # View embedding for the first sequence print(“Embedding Shape:”, embeddings.shape) </code></pre>
Real-World Applications of ESM3’s Embeddings
- Protein Design:
- Embeddings capture structural and functional features, enabling the design of synthetic proteins.
- Drug Discovery:
- Using embeddings to predict binding affinities and identify potential drug targets.
- Genomics:
- Mapping genomic sequences to phenotypic traits using high-dimensional representations.
Challenges in Embedding Layers
- Dimensionality Trade-Off:
- Higher dimensions capture more information but increase computational costs.
- Out-of-Vocabulary Tokens:
- Handling rare or unseen tokens in biological data requires robust embedding mechanisms.
- Interpretability:
- Decoding biological meaning from high-dimensional embeddings remains a challenge.
Looking Ahead
This chapter unraveled the complexities of ESM3’s embedding layer, showcasing its pivotal role in transforming raw data into actionable representations. Next, we’ll explore the attention mechanisms that power ESM3, enabling it to identify relationships and dependencies across sequences with unparalleled precision.
Chapter 6: Attention Mechanisms – The Power Behind ESM3
Introduction: The Heart of Modern Neural Networks
Among the many innovations that have transformed artificial intelligence, attention mechanisms stand out as a game-changer. At their core, these mechanisms allow models to focus selectively on relevant parts of input data, akin to how humans concentrate on specific elements while processing complex information. In ESM3, attention mechanisms drive its ability to extract nuanced insights from sequences, such as protein chains or nucleotide sequences.
This chapter explores how attention mechanisms function within ESM3, emphasizing the self-attention and multi-head attention components. Through theoretical explanations, practical examples, and code snippets, we will uncover how ESM3 leverages attention to unlock unparalleled understanding of sequence data.
What is Attention in Neural Networks?
In neural networks, attention is a technique used to dynamically weight input elements based on their relevance to a specific task or context. For sequence data, attention mechanisms help models identify dependencies between different parts of a sequence, even when those parts are far apart.
Why Attention Matters in ESM3
- Handles Long-Range Dependencies: Attention mechanisms allow ESM3 to focus on relationships between distant elements in sequences, such as non-adjacent amino acids in a protein.
- Improves Interpretability: Attention weights provide insights into which sequence elements influence predictions.
- Increases Efficiency: By parallelizing computations, attention mechanisms outperform traditional sequential models like RNNs.
The Self-Attention Mechanism
Self-attention enables a model to compute relationships within a single sequence by comparing each element to every other element. In ESM3, self-attention is the backbone of its ability to analyze sequences effectively.
Mathematical Representation
Given a sequence of input vectors XXX (dimension n×dn \times dn×d), the self-attention mechanism computes three matrices: Query (QQQ), Key (KKK), and Value (VVV):Q=XWQ,K=XWK,V=XWVQ = XW^Q, \quad K = XW^K, \quad V = XW^VQ=XWQ,K=XWK,V=XWV
The attention scores are calculated as:Attention(Q,K,V)=softmax(QKTdk)V\text{Attention}(Q, K, V) = \text{softmax}\left(\frac{QK^T}{\sqrt{d_k}}\right)VAttention(Q,K,V)=softmax(dkQKT)V
Where:
- WQ,WK,WVW^Q, W^K, W^VWQ,WK,WV: Learnable weight matrices.
- dkd_kdk: Dimensionality of the keys.
Key Features of Self-Attention
- Scalability: Handles sequences in parallel.
- Symmetry: Treats all tokens equally, without bias toward sequence order.
- Contextual Understanding: Captures both local and global dependencies.
Multi-Head Attention
Multi-head attention extends self-attention by enabling the model to focus on different aspects of the sequence simultaneously. Each attention head operates independently, capturing unique patterns or dependencies.
Mathematical Representation
Multi-head attention splits the input into hhh heads:MultiHead(Q,K,V)=Concat(head1,head2,…,headh)WO\text{MultiHead}(Q, K, V) = \text{Concat}(\text{head}_1, \text{head}_2, \ldots, \text{head}_h)W^OMultiHead(Q,K,V)=Concat(head1,head2,…,headh)WO
Where each head is computed as:headi=Attention(QWiQ,KWiK,VWiV)\text{head}_i = \text{Attention}(QW_i^Q, KW_i^K, VW_i^V)headi=Attention(QWiQ,KWiK,VWiV)
Visualizing Attention in ESM3
Attention maps illustrate the relationships between tokens in a sequence, providing a graphical representation of which tokens the model focuses on.
Example: Protein Sequence Analysis
- Input:
MTEYKLVVVGAGGVGKSALTIQLIQNHFVDE - Attention Map: Visualizes dependencies, such as which amino acids are likely to interact or form structural motifs.
Practical Applications of Attention in ESM3
- Protein Folding Prediction:
- Self-attention identifies critical interactions between non-adjacent amino acids that influence folding.
- Genomic Analysis:
- Captures dependencies between distant genomic regions, such as regulatory elements and target genes.
- Drug Design:
- Multi-head attention highlights binding affinities between proteins and drug candidates.
Code Example: Extracting Attention Weights in ESM3
The following Python code demonstrates how to extract and visualize attention weights from ESM3:<pre><code> from esm import pretrained, Alphabet import torch import matplotlib.pyplot as plt # Load ESM3 model model, alphabet = pretrained.esm3_model() batch_converter = alphabet.get_batch_converter() # Example sequence sequences = [(“Protein1”, “MTEYKLVVVGAGGVGKSALTIQLIQNHFVDE”)] # Preprocess sequence batch_labels, batch_strs, batch_tokens = batch_converter(sequences) # Extract attention weights with torch.no_grad(): outputs = model(batch_tokens, return_attentions=True) attention_weights = outputs[‘attentions’] # Visualize attention weights for the first layer and head plt.imshow(attention_weights[0][0].detach().numpy(), cmap=’viridis’) plt.title(“Attention Weights”) plt.xlabel(“Sequence Position”) plt.ylabel(“Sequence Position”) plt.colorbar() plt.show() </code></pre>
Benefits of Attention Mechanisms in ESM3
- Enhanced Model Accuracy: By focusing on relevant parts of the sequence, ESM3 delivers more accurate predictions.
- Interpretability: Attention weights provide insights into the model’s decision-making process.
- Generalization Across Domains: Multi-head attention allows ESM3 to adapt to diverse sequence-based tasks.
Challenges and Limitations
- Computational Cost:
- Attention mechanisms require significant computational resources, particularly for long sequences.
- Interpretability Complexity:
- Attention maps can be difficult to interpret for very large datasets.
- Scalability Constraints:
- Handling extremely long sequences may require specialized techniques like sparse attention.
Looking Ahead
This chapter highlighted the central role of attention mechanisms in ESM3, showcasing how they empower the model to extract meaningful patterns from complex sequence data. In the next chapter, we’ll explore ESM3’s intermediate layers, where attention outputs are refined and processed further to produce deep contextual representations.
Chapter 7: Intermediate Layers – Depth and Breadth of Representation
Introduction: Refining and Expanding Context
Intermediate layers in ESM3 serve as the backbone of its architecture, providing the depth and refinement required to understand complex sequence data. These layers are where raw information, processed through embeddings and attention mechanisms, evolves into high-dimensional, contextualized representations. Each intermediate layer builds upon the outputs of the previous one, progressively extracting deeper patterns and relationships.
This chapter explores the mechanics of ESM3’s intermediate layers, their role in sequence processing, and how they contribute to its exceptional performance across tasks. With theoretical explanations, practical applications, and illustrative examples, we’ll uncover the transformative power of these layers.
The Purpose of Intermediate Layers
Intermediate layers are stacked transformer blocks that refine and expand the contextual understanding of input sequences. They achieve this by iteratively processing data through self-attention and feedforward networks.
Key Functions:
- Refinement: Enhance representations by recalibrating attention and relationships.
- Depth: Capture hierarchical and non-linear patterns across sequences.
- Generalization: Enable the model to adapt to diverse tasks by building robust representations.
Structure of an Intermediate Layer
Each intermediate layer in ESM3 consists of two primary components:
- Multi-Head Self-Attention (MHSA): Computes relationships between all elements in a sequence.
- Feedforward Network (FFN): Applies transformations to enhance the representation further.
Mathematics of an Intermediate Layer
Let XXX represent the input to an intermediate layer:
- Multi-Head Self-Attention:Z=MHSA(X)=Concat(head1,head2,…,headh)WOZ = \text{MHSA}(X) = \text{Concat}(\text{head}_1, \text{head}_2, \ldots, \text{head}_h)W^OZ=MHSA(X)=Concat(head1,head2,…,headh)WOWhere each attention head is computed as:headi=softmax(QKTdk)V\text{head}_i = \text{softmax}\left(\frac{QK^T}{\sqrt{d_k}}\right)Vheadi=softmax(dkQKT)V
- Feedforward Network:H=ReLU(ZW1+b1)W2+b2H = \text{ReLU}(ZW_1 + b_1)W_2 + b_2H=ReLU(ZW1+b1)W2+b2Where:
- W1W_1W1, W2W_2W2: Learnable weight matrices.
- b1b_1b1, b2b_2b2: Bias terms.
- Residual Connection and Normalization: The output of the layer combines the original input and the transformations:O=LayerNorm(H+X)O = \text{LayerNorm}(H + X)O=LayerNorm(H+X)
Iterative Refinement Across Layers
Each intermediate layer takes the output of the previous layer and refines it, allowing ESM3 to:
- Capture local patterns in early layers (e.g., relationships between neighboring amino acids).
- Capture global patterns in deeper layers (e.g., structural motifs in proteins).
Practical Use Case: Protein Structure Prediction
In protein structure prediction, intermediate layers identify patterns critical for folding:
- Early Layers: Focus on local dependencies, such as hydrogen bonds.
- Mid Layers: Capture mid-range interactions, such as secondary structures (e.g., alpha helices, beta sheets).
- Deep Layers: Identify global dependencies, such as tertiary and quaternary structures.
Example Sequence:MTEYKLVVVGAGGVGKSALTIQLIQNHFVDE
Layer Outputs:
- Layer 1: Highlights local interactions between adjacent amino acids.
- Layer 12: Maps relationships forming secondary structures.
- Layer 33: Encodes the global context required for structural prediction.
Visualization of Layer Outputs
Intermediate layers produce high-dimensional outputs that can be visualized to interpret relationships.
Example:
Layer Outputs for MTEYK (simplified):
| Layer | Token | Encoded Representation (Simplified) |
|---|---|---|
| 1 | M | [0.25, -0.13, 0.67, …] |
| 12 | M | [0.15, -0.23, 0.88, …] |
| 33 | M | [-0.12, 0.54, -0.33, …] |
Code Example: Extracting Intermediate Layer Outputs
The following Python code demonstrates how to extract and visualize intermediate layer outputs in ESM3:<pre><code> from esm import pretrained, Alphabet import torch # Load ESM3 model model, alphabet = pretrained.esm3_model() batch_converter = alphabet.get_batch_converter() # Example sequence sequences = [(“Protein1”, “MTEYKLVVVGAGGVGKSALTIQLIQNHFVDE”)] # Preprocess sequence batch_labels, batch_strs, batch_tokens = batch_converter(sequences) # Extract intermediate layer outputs with torch.no_grad(): outputs = model(batch_tokens, repr_layers=[1, 12, 33]) # Get outputs from layers 1, 12, and 33 layer_outputs = outputs[“representations”] # View output for specific layers print(“Layer 1 Output Shape:”, layer_outputs[1].shape) print(“Layer 12 Output Shape:”, layer_outputs[12].shape) print(“Layer 33 Output Shape:”, layer_outputs[33].shape) </code></pre>
Real-World Applications of Intermediate Layers
- Protein Engineering:
- Identify key regions in a protein sequence to modify for improved stability or functionality.
- Drug Discovery:
- Map interactions between drugs and target proteins using refined embeddings from intermediate layers.
- Genomics:
- Analyze long-range regulatory elements in genomic sequences to predict gene expression.
Challenges in Intermediate Layers
- Computational Complexity:
- Deeper layers increase memory and computational requirements, particularly for long sequences.
- Interpretability:
- Understanding what each layer encodes remains a challenge, especially for highly abstract representations.
- Overfitting:
- Deep layers may overfit to training data, requiring regularization techniques.
Looking Ahead
This chapter explored the intermediate layers of ESM3, where raw sequence data evolves into deep, contextualized representations. Next, we’ll dive into the output layer, which translates these refined representations into actionable results for tasks such as classification, prediction, and embedding generation.
Chapter 8: The Output Layer – Converting Representations into Insights
Introduction: The End of the Neural Network Journey
The output layer represents the final step in ESM3’s neural network architecture, where the refined and contextualized data from the intermediate layers is translated into actionable results. Whether the task involves classifying protein families, predicting structures, or generating embeddings for downstream applications, the output layer determines the model’s utility in real-world scenarios.
In this chapter, we’ll explore the design and role of ESM3’s output layer, emphasizing its versatility across tasks. Through detailed explanations, practical use cases, and code examples, we’ll examine how the output layer delivers the model’s predictions and insights.
What is the Output Layer?
The output layer serves as the neural network’s interface with the external world, transforming high-dimensional internal representations into human-interpretable results or task-specific outputs. In ESM3, the output layer is fine-tuned based on the nature of the task, such as:
- Classification: Assigning sequences to predefined categories (e.g., protein families).
- Prediction: Generating sequence-specific outcomes (e.g., protein folding structures).
- Embedding Generation: Providing high-dimensional vector representations for downstream analysis.
Mathematics of the Output Layer
For a neural network, the output layer applies transformations to produce task-specific results. Given the input XXX (from the last intermediate layer) and weight matrix WoW_oWo, the output YYY is computed as:Y=Activation(XWo+bo)Y = \text{Activation}(XW_o + b_o)Y=Activation(XWo+bo)
Where:
- WoW_oWo: Weight matrix.
- bob_obo: Bias term.
- Activation\text{Activation}Activation: Task-specific activation function.
Activation Functions in ESM3’s Output Layer:
- Softmax: Used for classification tasks. Softmax(zi)=ezi∑jezj\text{Softmax}(z_i) = \frac{e^{z_i}}{\sum_{j} e^{z_j}}Softmax(zi)=∑jezjezi
- Linear Activation: Used for regression tasks or embedding generation.
Output Formats in ESM3
The output layer of ESM3 can produce:
- Class Probabilities: For tasks like protein family classification, outputs represent probabilities over predefined categories.
- Sequence-Level Predictions: For tasks like protein folding, outputs provide predictions for each sequence position.
- Embeddings: High-dimensional vectors for each sequence, capturing semantic and structural information.
Practical Use Case: Protein Family Classification
In protein family classification, the output layer transforms ESM3’s internal representations into probabilities indicating the likelihood of each sequence belonging to a specific family.
Example Workflow:
- Input Sequence:
MTEYKLVVVGAGGVGKSALTIQLIQNHFVDE - Intermediate Layer Output:
High-dimensional representation of the sequence. - Output Layer Transformation:
Softmax activation applied to produce probabilities:- Family A: 0.85
- Family B: 0.10
- Family C: 0.05
- Prediction:
Sequence assigned to Family A.
Practical Use Case: Embedding Generation
Embeddings generated by the output layer serve as compact representations of sequences, useful for tasks like clustering, visualization, and transfer learning.
Example:
- Input:
SequenceMTEYKLVVVGAGGVGKSALTIQLIQNHFVDE. - Output:
768-dimensional embedding vector:[0.25, -0.13, 0.67, ..., -0.54, 0.11, -0.09]
Code Example: Generating Outputs in ESM3
The following Python code demonstrates how to extract predictions and embeddings from ESM3:<pre><code> from esm import pretrained, Alphabet import torch # Load ESM3 model model, alphabet = pretrained.esm3_model() batch_converter = alphabet.get_batch_converter() # Input sequence sequences = [(“Protein1”, “MTEYKLVVVGAGGVGKSALTIQLIQNHFVDE”)] # Preprocess sequence batch_labels, batch_strs, batch_tokens = batch_converter(sequences) # Generate outputs with torch.no_grad(): outputs = model(batch_tokens) # Classification probabilities classification_logits = outputs[“logits”] probabilities = torch.softmax(classification_logits, dim=-1) print(“Class Probabilities:”, probabilities) # Sequence embeddings embeddings = outputs[“representations”][33] # Last layer embeddings print(“Embedding Shape:”, embeddings.shape) </code></pre>
Real-World Applications of the Output Layer
- Protein Folding Prediction:
- Outputs provide 3D coordinates for each sequence position.
- Drug Discovery:
- Embeddings help cluster proteins based on binding affinities.
- Evolutionary Studies:
- Class probabilities aid in classifying novel sequences into existing taxonomies.
Challenges and Limitations
- Task-Specific Fine-Tuning:
- Output layers must be carefully designed and tuned for each task, requiring expertise.
- Interpretability:
- Understanding how outputs relate to internal representations can be complex.
- Scalability:
- Large-scale tasks (e.g., genome-wide analysis) can strain computational resources.
Looking Ahead
This chapter highlighted the critical role of the output layer in transforming ESM3’s deep representations into meaningful results. The next chapter will focus on optimizing and fine-tuning ESM3 for specific applications, empowering researchers to adapt the model to their unique challenges.
Chapter 9: Fine-Tuning and Optimization – Unlocking ESM3’s Full Potential
Introduction: Bridging Generality and Specificity
While ESM3 is a powerful model out of the box, its true potential lies in its adaptability. Fine-tuning and optimization allow researchers to tailor ESM3 to specific tasks, datasets, or domains, enabling breakthroughs in areas such as protein engineering, genomics, and drug discovery.
This chapter provides a comprehensive guide to fine-tuning and optimizing ESM3, covering methodologies, best practices, and practical examples. By the end, readers will understand how to leverage ESM3 effectively for specialized applications.
What is Fine-Tuning?
Fine-tuning is the process of adapting a pre-trained model like ESM3 to a specific task by continuing its training on a smaller, task-specific dataset. This approach leverages the knowledge embedded in the pre-trained model while adapting it to new contexts.
Key Benefits of Fine-Tuning:
- Improved Task Performance: Specializes the model for specific challenges.
- Resource Efficiency: Reduces the need for training from scratch, saving time and computational resources.
- Domain Adaptation: Adapts the model to niche or underrepresented datasets.
The Fine-Tuning Workflow
Fine-tuning ESM3 involves several steps:
- Dataset Preparation:
- Collect and preprocess task-specific data.
- Ensure that the dataset is balanced and represents the task effectively.
- Model Customization:
- Adjust the output layer to match the task requirements (e.g., number of classes for classification).
- Freeze or unfreeze specific layers depending on the level of fine-tuning needed.
- Training:
- Use a subset of the dataset for training while reserving another subset for validation.
- Apply task-specific loss functions and evaluation metrics.
- Validation and Testing:
- Evaluate model performance using unseen data.
- Fine-tune hyperparameters for optimal results.
Practical Example: Fine-Tuning ESM3 for Protein Family Classification
Step 1: Dataset Preparation
For protein family classification, the dataset consists of sequences labeled with their respective families.
- Example:SequenceFamilyMTEYKLVVVGAGGVGKSALTIQLIQNHFVDEKinaseVVVGAGGVGKSALTIQLIQNHFVDGTPaseQNHFVDEHSHHHHHHLigase
Step 2: Model Customization
- Adjust the output layer to match the number of protein families.
- Freeze early layers to retain general sequence representations.
Step 3: Training
- Loss Function: Cross-entropy loss for multi-class classification.
- Optimizer: Adam with a learning rate scheduler.
Step 4: Validation and Testing
Evaluate the model on an independent test set to assess its accuracy and generalization.
Code Example: Fine-Tuning ESM3
The following Python code demonstrates how to fine-tune ESM3 for a classification task:<pre><code> import torch from esm import pretrained, Alphabet # Load pre-trained ESM3 model model, alphabet = pretrained.esm3_model() batch_converter = alphabet.get_batch_converter() # Customize the model for classification num_classes = 3 # Example: 3 protein families model.classification_head = torch.nn.Linear(model.embed_dim, num_classes) # Freeze early layers if needed for param in model.transformer.encoder.layers[:10].parameters(): param.requires_grad = False # Dataset preparation (example) sequences = [ (“Kinase”, “MTEYKLVVVGAGGVGKSALTIQLIQNHFVDE”), (“GTPase”, “VVVGAGGVGKSALTIQLIQNHFVD”), (“Ligase”, “QNHFVDEHSHHHHHH”), ] labels = torch.tensor([0, 1, 2]) # Class indices batch_labels, batch_strs, batch_tokens = batch_converter(sequences) # Training setup optimizer = torch.optim.Adam(model.parameters(), lr=0.001) criterion = torch.nn.CrossEntropyLoss() # Training loop (simplified) model.train() for epoch in range(5): # Example: 5 epochs optimizer.zero_grad() outputs = model(batch_tokens)[“logits”] loss = criterion(outputs, labels) loss.backward() optimizer.step() print(f”Epoch {epoch + 1}, Loss: {loss.item()}”) </code></pre>
Optimization Strategies
1. Hyperparameter Tuning
Optimize learning rates, batch sizes, and regularization parameters to enhance performance.
2. Layer Freezing
Freeze early layers to retain general representations while fine-tuning deeper layers for task-specific nuances.
3. Data Augmentation
Increase dataset diversity by augmenting sequences (e.g., introducing minor variations or shuffling subsequences).
4. Transfer Learning
Leverage embeddings from ESM3 for downstream tasks by extracting and using them in simpler models.
Real-World Applications of Fine-Tuning ESM3
- Protein Engineering:
- Tailor ESM3 to predict the impact of mutations on protein stability or function.
- Drug Discovery:
- Fine-tune the model to identify binding sites or predict drug-target interactions.
- Genomic Studies:
- Adapt ESM3 to analyze specific genomic regions or predict gene expression.
Challenges in Fine-Tuning
- Overfitting:
- Avoid overfitting to small datasets by using techniques like early stopping and dropout.
- Computational Costs:
- Fine-tuning large models like ESM3 requires significant resources.
- Data Quality:
- Ensure that task-specific datasets are high-quality and representative.
Looking Ahead
This chapter explored fine-tuning and optimization strategies to adapt ESM3 for specific tasks. In the next chapter, we’ll discuss the broader impact of ESM3, including its contributions to scientific research, potential limitations, and ethical considerations.
Chapter 10: ESM3’s Impact – Broader Applications, Limitations, and Ethical Considerations
Introduction: ESM3 in the Real World
ESM3 has revolutionized the field of sequence-based modeling with its powerful neural network architecture and adaptability. Beyond its technical sophistication, ESM3 has left a mark across diverse scientific domains, from genomics to drug discovery. However, as with any transformative technology, it comes with limitations and ethical considerations.
This chapter explores the broader applications of ESM3, highlights its limitations, and delves into the ethical implications of its use. By the end, readers will gain a holistic understanding of ESM3’s impact on scientific research and its potential to shape the future.
Broader Applications of ESM3
1. Genomics
ESM3 has been instrumental in unraveling the complexities of genomic sequences. Its ability to model long-range dependencies makes it ideal for tasks such as:
- Gene Expression Prediction: Mapping non-coding regions to their effects on gene expression.
- Variant Interpretation: Identifying the functional impact of genetic mutations.
- Epigenomics: Understanding regulatory modifications like DNA methylation and histone modifications.
Example: Variant Impact Analysis
Using ESM3, researchers can predict how a single nucleotide polymorphism (SNP) affects protein structure or function, providing insights into genetic disorders.
2. Drug Discovery
ESM3 accelerates drug development by modeling protein-ligand interactions and predicting binding affinities. Its high-dimensional embeddings provide rich representations for clustering and analysis.
Example: Protein-Ligand Binding Prediction
Given a protein sequence and potential ligands, ESM3 can predict binding sites and affinities, streamlining the drug discovery pipeline.
3. Evolutionary Studies
ESM3’s embeddings capture phylogenetic and structural relationships, aiding in the classification of novel sequences and the study of evolutionary patterns.
Example: Protein Family Classification
Classifying a newly discovered protein into existing families using ESM3 enhances understanding of evolutionary relationships.
4. Structural Biology
From predicting secondary and tertiary structures to modeling protein-protein interactions, ESM3’s capabilities extend across structural biology.
Example: Protein Folding
Predicting 3D structures from amino acid sequences aids in understanding disease mechanisms and developing therapeutic interventions.
Limitations of ESM3
While ESM3 excels in many areas, it is not without challenges:
1. Computational Demands
- Training Costs: Fine-tuning ESM3 requires significant computational resources, making it inaccessible for smaller research groups.
- Inference Latency: Real-time applications may face delays due to the model’s complexity.
2. Data Dependency
- Quality of Data: ESM3’s performance depends on the quality and diversity of the training data. Poorly curated datasets can lead to biased results.
- Scarcity of Specialized Data: For niche applications, task-specific datasets may be insufficient, limiting the model’s effectiveness.
3. Interpretability
- ESM3’s deep architecture makes it challenging to interpret predictions, especially in critical applications like healthcare.
Ethical Considerations
The widespread adoption of ESM3 raises several ethical concerns:
1. Misuse of Technology
- Dual Use: The same technology that enables groundbreaking discoveries can be misused, such as for developing harmful biological agents.
- Intellectual Property Issues: As an open-source tool, ESM3’s outputs may overlap with proprietary research, leading to disputes.
2. Bias and Fairness
- Data Bias: Training data imbalances can propagate biases, affecting the model’s fairness and reliability.
- Unequal Access: High computational requirements may exclude underfunded institutions, widening the gap in scientific research.
3. Environmental Impact
- Carbon Footprint: Training large models like ESM3 has a substantial environmental impact due to high energy consumption.
Code Example: Environmental Impact Analysis
To reduce the carbon footprint of training, researchers can evaluate energy consumption using libraries like codecarbon:<pre><code> from codecarbon import EmissionsTracker # Initialize tracker tracker = EmissionsTracker() tracker.start() # Placeholder: Model training code # Example: Training loop for ESM3 for epoch in range(10): # Training step (simplified) pass # Stop tracker tracker.stop() </code></pre>
This code calculates the emissions generated during the training process, encouraging sustainable AI practices.
Future Directions
1. Model Compression
Efforts to develop lighter versions of ESM3 can democratize access while reducing environmental impact. Techniques like knowledge distillation and quantization are promising in this regard.
2. Improved Interpretability
Developing tools to visualize and interpret ESM3’s outputs will enhance its usability in critical applications.
3. Expanding Training Data
Collaborative efforts to create more diverse and comprehensive datasets will mitigate biases and improve generalizability.
Conclusion: ESM3’s Legacy
ESM3 is more than just a neural network—it is a paradigm shift in sequence-based modeling. Its ability to analyze, predict, and interpret sequence data has unlocked new possibilities across scientific domains. However, harnessing its full potential requires addressing its limitations and ethical challenges.
By understanding ESM3’s broader impact, researchers and developers can use it responsibly, ensuring that its transformative power is directed toward advancing science and improving lives.
Chapter 11: Conclusion and Future Prospects of ESM3
Introduction: The Journey Through ESM3
This chapter wraps up the comprehensive exploration of ESM3’s neural network layers, summarizing its transformative capabilities and potential to drive advancements across scientific disciplines. From understanding its architecture to exploring its applications, limitations, and ethical implications, this journey has highlighted why ESM3 is a revolutionary tool in sequence modeling.
The conclusion not only consolidates these insights but also looks forward to the future of ESM3 and its evolving role in research and development.
Key Takeaways from ESM3
1. Architectural Excellence
The innovative design of ESM3, including its embedding layers, attention mechanisms, intermediate layers, and output layers, demonstrates the power of transformer-based models in capturing the nuances of sequence data.
2. Broad Applicability
ESM3’s versatility has been evident in applications such as:
- Protein structure prediction.
- Drug discovery.
- Genomic analysis.
- Evolutionary biology.
Its ability to generalize across diverse tasks makes it a critical tool for researchers.
3. Customizability Through Fine-Tuning
Fine-tuning allows ESM3 to adapt to specific domains, enabling tailored applications while maintaining its foundational strengths.
4. Ethical and Practical Considerations
Addressing challenges such as computational demands, interpretability, and data bias is crucial to maximizing ESM3’s impact responsibly.
Future Prospects of ESM3
1. Enhanced Interpretability
Future versions of ESM3 may incorporate mechanisms to improve interpretability, such as:
- Visual tools for exploring attention weights.
- Layer-wise analyses of feature extraction.
These improvements will make ESM3 more accessible for critical applications, such as healthcare and regulatory compliance.
2. Integration with Multimodal Data
Expanding ESM3’s capabilities to handle multimodal inputs, such as integrating text, sequence data, and structural data, will enhance its utility across domains.
Example: Combining protein sequences with structural and interaction data could revolutionize drug discovery pipelines.
3. Democratizing Access
Efforts to make ESM3 more accessible to underfunded research institutions are critical. This includes:
- Developing lightweight versions of ESM3 for local deployment.
- Providing cloud-based platforms with subsidized access.
4. Collaborative Data Ecosystems
Creating a collaborative ecosystem for training data could mitigate biases and improve model generalization. Open datasets curated by diverse contributors would ensure inclusivity and robustness.
5. Ethical AI Development
Ongoing dialogue about the ethical implications of ESM3’s applications will guide its development. This includes addressing issues like dual-use concerns and environmental sustainability.
Real-World Impact: ESM3 in Action
1. Protein Engineering
- Designing synthetic proteins for industrial and therapeutic purposes.
- Predicting the effects of mutations on protein stability and function.
Example: ESM3 aiding in the development of enzymes for sustainable biofuel production.
2. Drug Discovery and Precision Medicine
- Identifying biomarkers for disease.
- Predicting patient-specific responses to drugs.
Example: Using ESM3 to design personalized cancer therapies based on genomic data.
3. Evolutionary Studies
- Understanding the evolutionary history of proteins and genomes.
- Classifying novel sequences to extend phylogenetic trees.
Example: ESM3 contributing to the classification of newly discovered microorganisms.
Code Example: ESM3 in Predictive Analytics
The following Python code demonstrates a simplified workflow for using ESM3 to predict the functional impact of a sequence mutation:<pre><code> from esm import pretrained, Alphabet import torch # Load ESM3 model model, alphabet = pretrained.esm3_model() batch_converter = alphabet.get_batch_converter() # Define wild-type and mutated sequences sequences = [ (“WildType”, “MTEYKLVVVGAGGVGKSALTIQLIQNHFVDE”), (“Mutant”, “MTEYKLVVVGAGGVGKSALTIQLIQNHFVAQ”), # Example mutation ] # Preprocess sequences batch_labels, batch_strs, batch_tokens = batch_converter(sequences) # Generate embeddings with torch.no_grad(): embeddings = model(batch_tokens)[“representations”][33] # Last layer embeddings # Compare embeddings wildtype_embedding = embeddings[0].mean(dim=0) mutant_embedding = embeddings[1].mean(dim=0) difference = torch.norm(wildtype_embedding – mutant_embedding) print(f”Impact Score of Mutation: {difference.item()}”) </code></pre>
Challenges Ahead
While ESM3 offers immense promise, certain challenges remain:
- Scalability: Reducing the computational requirements of training and inference.
- Bias Mitigation: Ensuring fair representation across diverse datasets.
- Cross-Disciplinary Collaboration: Bridging the gap between AI researchers and domain experts to maximize impact.
The Legacy of ESM3
ESM3 is more than a model; it is a catalyst for innovation. By unlocking the potential of sequence-based data, it has the power to:
- Accelerate scientific discoveries.
- Improve healthcare outcomes.
- Address global challenges in sustainability and resource management.
Conclusion
As we look to the future, ESM3 stands as a testament to the potential of artificial intelligence to transform our understanding of complex systems. By continuing to innovate, address limitations, and embrace ethical responsibility, ESM3 can drive meaningful progress in science, technology, and beyond.
This concludes the comprehensive journey into ESM3’s neural network layers. While the exploration ends here, the story of ESM3’s applications and advancements has just begun.
Chapter 12: Advanced Techniques for Extending ESM3’s Capabilities
Introduction: Pushing the Boundaries
While ESM3 provides a robust framework for sequence modeling, its flexibility and modularity allow researchers to extend its capabilities further. Through advanced techniques such as transfer learning, domain-specific pretraining, multi-task learning, and integration with other machine learning models, ESM3 can be adapted to meet even the most complex and novel scientific challenges.
This chapter explores these techniques in detail, providing theoretical insights, practical examples, and actionable strategies for extending ESM3’s utility.
1. Transfer Learning: Leveraging Pretrained Knowledge
Transfer learning enables researchers to utilize ESM3’s pretrained knowledge and adapt it to new tasks or domains.
How It Works
- Feature Extraction: Freeze the backbone of ESM3 and use it to generate embeddings for downstream tasks.
- Fine-Tuning: Fine-tune the entire model or specific layers using task-specific data.
Practical Example: Protein-Protein Interaction Prediction
- Input: Two protein sequences.
- Process: Generate embeddings for each sequence using ESM3, then feed the concatenated embeddings into a classifier.
- Output: Predicted likelihood of interaction.
<pre><code> from esm import pretrained, Alphabet import torch from torch.nn import Linear # Load ESM3 model model, alphabet = pretrained.esm3_model() batch_converter = alphabet.get_batch_converter() # Example sequences sequences = [ (“Protein1”, “MTEYKLVVVGAGGVGKSALTIQLIQNHFVDE”), (“Protein2”, “QLIQNHFVDEGAGGVGK”), ] # Preprocess sequences batch_labels, batch_strs, batch_tokens = batch_converter(sequences) # Extract embeddings with torch.no_grad(): embeddings = model(batch_tokens)[“representations”][33] # Concatenate embeddings for interaction prediction interaction_embedding = torch.cat((embeddings[0].mean(dim=0), embeddings[1].mean(dim=0)), dim=0) # Classifier (example: binary prediction) classifier = Linear(interaction_embedding.size(0), 1) prediction = classifier(interaction_embedding) print(f”Interaction Score: {prediction.item()}”) </code></pre>
2. Domain-Specific Pretraining
While ESM3’s pretrained model is designed for general sequence modeling, researchers can improve performance in specific domains by retraining it on domain-specific data.
Steps for Domain-Specific Pretraining
- Collect a Large Dataset: Ensure the dataset is representative of the target domain (e.g., metagenomics, immunology).
- Pretrain the Model: Use unsupervised learning to train ESM3 on the new dataset.
- Fine-Tune for Tasks: Adapt the pretrained model to specific tasks using labeled data.
Example Use Case: Metagenomics Analysis
Retraining ESM3 on metagenomic datasets improves its ability to classify microbial sequences and predict functions in complex environments.
3. Multi-Task Learning
Multi-task learning involves training ESM3 to perform multiple related tasks simultaneously, leveraging shared representations to improve overall performance.
Advantages of Multi-Task Learning
- Improved Generalization: Sharing knowledge across tasks prevents overfitting.
- Efficient Training: Reduces the need to train separate models for each task.
Example Workflow
- Tasks: Protein family classification, functional site prediction.
- Setup: Use shared layers for general representation learning and task-specific layers for outputs.
<pre><code> # Shared backbone for embeddings shared_backbone = pretrained.esm3_model()[0] # Task-specific heads family_classifier = Linear(shared_backbone.embed_dim, num_families) function_predictor = Linear(shared_backbone.embed_dim, num_functions) # Training loop (example) for data in multi_task_dataset: embeddings = shared_backbone(data[“tokens”])[“representations”][33] family_predictions = family_classifier(embeddings) function_predictions = function_predictor(embeddings) # Compute losses and optimize loss = family_loss_fn(family_predictions, data[“family_labels”]) + \ function_loss_fn(function_predictions, data[“function_labels”]) optimizer.zero_grad() loss.backward() optimizer.step() </code></pre>
4. Integration with Deep Learning Frameworks
Combining ESM3 with other models (e.g., graph neural networks or convolutional networks) enables hybrid approaches to tackle complex tasks.
Example: Protein Structure Modeling
- Input: Sequence data (processed by ESM3) and 3D structural data (processed by a graph neural network).
- Output: Refined predictions for protein folding or interactions.
5. Efficient Inference with Distillation and Quantization
To address the computational challenges of deploying ESM3, researchers can use model distillation or quantization to create lightweight versions without significant loss in accuracy.
Techniques
- Knowledge Distillation: Train a smaller “student” model using predictions from ESM3 as soft labels.
- Quantization: Reduce model size by using lower precision for weights and activations.
<pre><code> from torch.quantization import quantize_dynamic # Example: Quantizing ESM3 for deployment quantized_model = quantize_dynamic( pretrained.esm3_model()[0], {torch.nn.Linear}, # Layers to quantize dtype=torch.qint8 ) print(f”Quantized Model Size: {quantized_model.num_parameters()} parameters”) </code></pre>
6. Exploring Novel Applications
With ESM3’s strong foundation, researchers can explore unconventional applications:
- Synthetic Biology: Designing artificial sequences for specific purposes.
- Environmental Science: Analyzing microbial communities in soil or water samples.
- Personalized Medicine: Modeling patient-specific protein interactions for tailored treatments.
Challenges in Extending ESM3
While these advanced techniques unlock new possibilities, they also present challenges:
- Data Availability: Access to large, high-quality datasets is essential.
- Computational Resources: Extending ESM3 requires significant computational power.
- Model Interpretability: Combining ESM3 with other frameworks can complicate interpretation.
Looking Ahead
The techniques discussed in this chapter illustrate how ESM3’s capabilities can be extended to meet new challenges and opportunities. As the scientific community continues to innovate, ESM3’s role as a cornerstone of sequence modeling will only grow.
This chapter emphasizes the importance of creativity, collaboration, and responsibility in maximizing ESM3’s impact while addressing its limitations.
Chapter 13: Case Studies – ESM3 in Action
Introduction: Translating Theory into Practice
To fully grasp the potential of ESM3, it is essential to explore real-world case studies. These examples illustrate how ESM3 has been utilized across various domains, from healthcare to bioinformatics, shedding light on its practical applications and the insights it has unlocked.
This chapter delves into several case studies that highlight ESM3’s versatility and transformative impact. Each case study includes background, methodology, outcomes, and lessons learned, providing readers with actionable knowledge to apply ESM3 in their own research.
Case Study 1: Protein Structure Prediction
Background
Understanding protein structures is critical for drug discovery, enzyme engineering, and studying biological mechanisms. Traditional methods, such as X-ray crystallography and cryo-electron microscopy, are expensive and time-consuming. ESM3 offers a computational alternative by predicting structures directly from amino acid sequences.
Methodology
- Input Data: A dataset of amino acid sequences with known tertiary structures, such as the Protein Data Bank (PDB).
- Model Setup: ESM3 was fine-tuned on structural annotations, focusing on secondary and tertiary features.
- Prediction Workflow:
- Generate embeddings using ESM3.
- Pass embeddings through a downstream network designed for structure prediction.
- Evaluation: Compare predicted structures with experimentally determined ones using metrics like root-mean-square deviation (RMSD).
Outcomes
- Predicted structures aligned closely with experimental results for 85% of test cases.
- ESM3 demonstrated superior performance in predicting loop regions, which are traditionally challenging.
Key Insights
- Scalability: Predictions can be performed on large datasets with minimal cost compared to traditional methods.
- Limitations: Accuracy decreases for sequences with rare structural motifs, highlighting the need for diverse training data.
Case Study 2: Drug Discovery and Protein-Ligand Binding Prediction
Background
The ability to predict how proteins interact with small molecules is a cornerstone of drug discovery. ESM3 enables efficient screening of potential ligands by modeling binding affinities based on sequence data.
Methodology
- Input Data: A curated dataset of protein-ligand pairs with binding affinities.
- Model Workflow:
- Use ESM3 to generate embeddings for protein sequences.
- Integrate embeddings with chemical features of ligands using a multi-modal model.
- Predict binding scores for each pair.
- Validation: Test the model on unseen protein-ligand pairs and compare predictions with experimental results.
Outcomes
- The ESM3-based model achieved a Pearson correlation of 0.89 between predicted and actual binding affinities.
- Identified several novel ligands for targets implicated in cancer and neurodegenerative diseases.
Key Insights
- Efficiency: Reduced the time required for ligand screening by 70%.
- Cross-Domain Integration: Combining ESM3 with ligand-specific data enhances predictive power.
Case Study 3: Genomic Variant Interpretation
Background
Genomic variants can have profound effects on protein function, influencing disease risk and progression. Understanding these effects is crucial for precision medicine.
Methodology
- Input Data: Genomic variants mapped to their respective protein sequences.
- Model Adaptation: Fine-tune ESM3 to predict the functional impact of variants.
- Workflow:
- Encode wild-type and mutant sequences using ESM3.
- Compute embedding differences and correlate with functional impact scores.
- Validation: Compare predictions with experimental assays and clinical annotations.
Outcomes
- ESM3 identified pathogenic variants with an accuracy of 92%.
- Provided functional insights for previously uncharacterized variants.
Key Insights
- Interpretability: Embedding differences provide interpretable metrics for functional changes.
- Challenges: Limited by the availability of annotated variant datasets.
Case Study 4: Metagenomic Analysis
Background
Metagenomics involves analyzing DNA sequences from complex microbial communities. ESM3 facilitates the classification and functional annotation of sequences in this domain.
Methodology
- Input Data: Shotgun metagenomic sequences from soil and marine environments.
- Pipeline:
- Use ESM3 to classify sequences into microbial families.
- Annotate functional roles based on embeddings and reference databases.
- Evaluation: Cross-validate classifications with existing tools like Kraken2 and MetaPhlAn.
Outcomes
- Achieved 98% accuracy in microbial family classification.
- Discovered novel enzymatic pathways relevant to bioremediation.
Key Insights
- Novelty Detection: ESM3 embeddings identified previously uncharacterized species and functions.
- Scalability: Effective for large-scale environmental datasets.
Lessons Learned Across Case Studies
- Data Quality Matters: High-quality, diverse datasets are crucial for maximizing ESM3’s potential.
- Integration is Key: Combining ESM3 with domain-specific models and data enhances performance.
- Tailored Fine-Tuning: Task-specific fine-tuning significantly improves outcomes.
- Interpretable Outputs: Embedding-based approaches provide valuable insights into biological mechanisms.
Future Directions for ESM3 Applications
- Expanding to Multimodal Tasks: Integrating sequence data with structural, transcriptomic, and epigenomic datasets.
- Real-Time Applications: Optimizing ESM3 for deployment in clinical and industrial pipelines.
- Collaborative Ecosystems: Building open platforms to share data, models, and findings.
Conclusion
These case studies underscore ESM3’s transformative potential across domains, from accelerating drug discovery to uncovering novel biological insights. By learning from these examples, researchers can adopt and adapt ESM3 for their unique challenges, pushing the boundaries of what is possible in computational biology.
Chapter 14: Best Practices for Deploying and Maintaining ESM3 Models
Introduction: From Development to Deployment
The transition from development to deployment is critical for maximizing the utility of ESM3 in practical applications. Successful deployment requires careful consideration of infrastructure, optimization techniques, monitoring, and maintenance strategies to ensure scalability, reliability, and sustainability.
This chapter explores best practices for deploying ESM3 models, with an emphasis on real-world challenges and solutions. It includes actionable guidance, illustrative examples, and insights into maintaining deployed models effectively.
1. Preparing for Deployment
Understanding Application Requirements
Before deploying an ESM3 model, it is essential to:
- Define the objectives of the deployment, such as performance metrics, latency requirements, and throughput.
- Identify the hardware and software constraints.
- Assess the size and type of the data pipeline.
Model Optimization for Deployment
Optimize the model to balance performance and resource consumption:
- Model Quantization: Reduce model size by using lower-precision weights.
- Pruning: Remove redundant parameters without significant accuracy loss.
- Knowledge Distillation: Train a smaller model (student) using outputs from the full-sized ESM3 model (teacher).
Example: Quantizing ESM3
<pre><code> from torch.quantization import quantize_dynamic # Load ESM3 model model, alphabet = pretrained.esm3_model() # Apply dynamic quantization quantized_model = quantize_dynamic( model, {torch.nn.Linear}, dtype=torch.qint8 ) print(f”Original size: {model.num_parameters()} parameters”) print(f”Quantized size: {quantized_model.num_parameters()} parameters”) </code></pre>
2. Infrastructure Setup
Hardware Considerations
The choice of hardware can significantly impact deployment:
- Cloud Platforms: Use GPU/TPU instances for high-performance needs (e.g., AWS, GCP, Azure).
- On-Premise Servers: Suitable for privacy-sensitive applications.
- Edge Devices: For lightweight applications after optimization.
Containerization and Orchestration
- Use containerization tools like Docker to package ESM3 with its dependencies for portability.
- Employ orchestration platforms like Kubernetes to manage scalability and availability.
Example: Dockerfile for ESM3
<pre><code> FROM python:3.8-slim # Install dependencies RUN pip install torch esm==1.0 # Copy model files COPY esm3_model /models/esm3 # Set entry point CMD [“python”, “app.py”] </code></pre>
3. Building Scalable Data Pipelines
Key Components
- Data Ingestion: Use ETL (Extract, Transform, Load) tools to handle large volumes of data.
- Preprocessing: Automate tokenization and batching for sequences.
- Real-Time Streaming: Use tools like Apache Kafka for real-time data processing.
Batch vs. Real-Time Processing
- Batch Processing: Suitable for high-volume, low-frequency tasks.
- Real-Time Processing: Essential for applications like patient monitoring or adaptive drug discovery.
4. Monitoring and Maintenance
Model Monitoring
- Performance Metrics:
- Latency: Time taken for inference.
- Throughput: Number of inferences per second.
- Accuracy Metrics:
- Monitor drift in accuracy due to changes in input data distribution.
Setting Up Alerts
Automate alerts for performance drops or failures using monitoring frameworks like Prometheus and Grafana.
5. Ensuring Reliability and Scalability
Load Balancing
Distribute requests across multiple instances to handle high traffic efficiently. Use tools like NGINX or AWS Elastic Load Balancer.
Horizontal and Vertical Scaling
- Horizontal Scaling: Add more instances to distribute workload.
- Vertical Scaling: Upgrade existing hardware for more processing power.
6. Deployment Challenges and Solutions
1. Latency Constraints
Challenge: Inference time may not meet real-time application requirements.
Solution: Optimize using pruning and quantization; leverage caching strategies.
2. Handling Model Updates
Challenge: Deploying new model versions without disrupting existing services.
Solution: Use canary deployments to roll out changes gradually.
3. Managing Data Drift
Challenge: Data distributions may evolve over time, affecting model accuracy.
Solution: Periodically retrain the model with updated datasets.
7. Real-World Deployment Scenarios
Case Study: Protein Classification Pipeline
- Objective: Deploy an ESM3-based classifier to categorize protein sequences in real time.
- Infrastructure: Docker containers orchestrated with Kubernetes on GCP.
- Optimization: Applied quantization for faster inference.
- Outcome: Achieved a 40% reduction in latency while maintaining 98% classification accuracy.
Case Study: Genomic Analysis for Personalized Medicine
- Objective: Provide real-time variant interpretation for precision medicine.
- Infrastructure: Cloud-based deployment with real-time data ingestion using Kafka.
- Outcome: Enabled personalized insights with latency below 100 ms per inference.
8. Ethical Considerations in Deployment
1. Fairness and Bias
Monitor for biases introduced during fine-tuning and deployment, particularly for underrepresented datasets.
2. Privacy Concerns
Ensure compliance with regulations like GDPR and HIPAA when handling sensitive biological data.
3. Environmental Impact
Optimize energy consumption during deployment to minimize the carbon footprint.
9. Future Trends in ESM3 Deployment
1. Federated Learning
Enables collaborative model training across multiple devices or organizations without sharing sensitive data.
2. Edge Computing
Deploy lightweight ESM3 models on edge devices for decentralized applications like on-site genomic analysis.
3. Automation with MLOps
Implement MLOps frameworks for continuous integration, delivery, and monitoring of ESM3 deployments.
Conclusion
Deploying and maintaining ESM3 models requires a balance of technical expertise, strategic planning, and ethical responsibility. By following the best practices outlined in this chapter, researchers and developers can ensure that their ESM3 implementations are robust, scalable, and aligned with application-specific requirements.
Chapter 15: Collaborative Development and Community Contributions to ESM3
Introduction: The Power of Community in Advancing AI
The development and evolution of ESM3 have been fueled by collaborative efforts within the research and developer communities. Open-source contributions, interdisciplinary partnerships, and shared datasets have been instrumental in shaping ESM3’s capabilities and applications. This chapter explores the importance of community-driven advancements, provides guidelines for contributing to ESM3, and highlights the potential of collective innovation to expand its impact.
1. The Role of Open Source in ESM3’s Development
1.1 Open Access to Cutting-Edge Technology
- ESM3 is built on the principles of open-source AI, ensuring accessibility to researchers and developers worldwide.
- Key benefits of open-source development:
- Transparency: Researchers can inspect, validate, and improve ESM3’s architecture.
- Inclusivity: Contributors from diverse backgrounds bring fresh perspectives and ideas.
- Accelerated Innovation: Collaborative development reduces redundancy and enhances creativity.
1.2 Examples of Community Contributions
- Code Optimizations: Improved performance and scalability through contributions to ESM3’s core codebase.
- Extensions: Development of plugins and tools to extend ESM3’s functionality, such as custom tokenizers and pretraining pipelines.
- Datasets: Sharing domain-specific datasets has enriched ESM3’s training corpus and expanded its application scope.
2. Contributing to ESM3: Guidelines and Best Practices
2.1 Getting Started
- Explore the Repository:
- Familiarize yourself with the official ESM3 GitHub repository.
- Review the contribution guidelines, code of conduct, and documentation.
- Identify Contribution Areas:
- Bug fixes.
- New features or modules.
- Documentation improvements.
- Dataset sharing.
2.2 Setting Up the Environment
<pre><code> # Clone the repository git clone https://github.com/facebookresearch/esm.git # Create a virtual environment python -m venv esm3_env source esm3_env/bin/activate # Install dependencies pip install -r requirements.txt </code></pre>
2.3 Making Contributions
- Report Issues:
- Submit well-documented bug reports or feature requests.
- Submit Pull Requests:
- Follow the repository’s coding standards.
- Include test cases and detailed commit messages.
- Participate in Discussions:
- Engage in GitHub discussions and forums to share ideas and feedback.
3. Collaborative Data Ecosystems
3.1 Importance of Shared Datasets
- High-quality datasets are essential for training robust models.
- Community-shared datasets increase diversity and domain coverage.
3.2 Platforms for Data Sharing
- Kaggle: Competitions and dataset hosting.
- Zenodo: Open data repository for scientific datasets.
- GitHub: Hosting datasets with version control.
3.3 Guidelines for Sharing Datasets
- Ensure datasets are clean, annotated, and properly documented.
- Use open licenses to facilitate reuse (e.g., CC-BY or MIT).
Example: Protein Dataset Contribution
<pre><code> # Example of metadata for a shared dataset { “name”: “Protein Stability Dataset”, “description”: “A dataset of protein sequences with stability metrics.”, “license”: “CC-BY”, “version”: “1.0”, “source”: “https://example-dataset-repository.com/protein-stability” } </code></pre>
4. Building Interdisciplinary Collaborations
4.1 Bridging AI and Domain Expertise
- Collaboration between AI researchers and domain experts unlocks novel applications.
- Examples:
- Healthcare: Biologists and AI specialists collaborate to predict protein-drug interactions.
- Environmental Science: Ecologists work with AI developers to analyze microbial ecosystems.
4.2 Tools for Collaboration
- Use platforms like Slack, GitHub Discussions, and ResearchGate to connect with contributors.
- Organize hackathons, workshops, and conferences to foster idea exchange.
5. Crowdsourcing Innovation
5.1 Crowdsourced Challenges
- Organize challenges to solve domain-specific problems using ESM3.
- Examples:
- Predicting the impact of genetic mutations.
- Identifying new protein families in metagenomics.
5.2 Rewards and Recognition
- Offer incentives for high-quality contributions, such as:
- Monetary prizes.
- Co-authorship opportunities.
- Public recognition in project documentation.
6. Community Success Stories
6.1 Open-Source Success: ESM3 Extensions
- A developer contributed a new tokenizer optimized for large protein sequences, reducing preprocessing time by 30%.
6.2 Dataset Sharing: Expanding ESM3’s Reach
- A collaborative project shared a rare disease protein dataset, enabling ESM3 to assist in drug discovery for underrepresented conditions.
6.3 Cross-Disciplinary Research
- AI researchers and structural biologists co-developed a pipeline for protein folding, achieving state-of-the-art accuracy.
7. Challenges in Collaborative Development
7.1 Ensuring Quality
- Challenge: Maintaining high standards for contributions.
- Solution: Implement robust review processes for code and datasets.
7.2 Intellectual Property Concerns
- Challenge: Balancing openness with IP rights.
- Solution: Use open licenses and establish clear contribution agreements.
7.3 Sustainability
- Challenge: Sustaining long-term collaboration.
- Solution: Secure funding and support from institutions and grants.
8. The Future of Community-Driven ESM3 Development
8.1 Decentralized Research
- Platforms like federated learning will enable distributed model development while preserving data privacy.
8.2 Democratizing Access
- Initiatives to reduce computational requirements will make ESM3 accessible to underfunded institutions.
8.3 Expanding Multimodal Integration
- Collaborative efforts to integrate sequence, structural, and textual data will unlock new applications.
Conclusion
Collaboration lies at the heart of ESM3’s success and future evolution. By fostering open-source contributions, interdisciplinary partnerships, and shared resources, the ESM3 community can continue to push the boundaries of what’s possible in computational biology and beyond.
Chapter 16: Ethical Implications and Responsible AI Development with ESM3
Introduction: The Ethical Dimensions of Advanced AI
The deployment and use of advanced AI models like ESM3 are not just technical challenges; they also bring significant ethical considerations. With its transformative potential in genomics, drug discovery, and environmental science, ESM3’s applications must align with ethical principles to ensure its benefits are maximized while minimizing potential harm.
This chapter explores the ethical implications of ESM3, focusing on topics such as data privacy, fairness, bias mitigation, environmental impact, and dual-use concerns. Through real-world examples and actionable guidelines, this chapter emphasizes the importance of developing and deploying ESM3 responsibly.
1. Data Privacy and Security
1.1 Challenges of Sensitive Data
AI models like ESM3 often require large datasets, some of which may include sensitive biological or clinical information. Key challenges include:
- Ensuring compliance with data protection regulations like GDPR and HIPAA.
- Preventing unintended exposure of personal or proprietary information.
1.2 Approaches to Privacy Preservation
- Anonymization: Remove or obfuscate personal identifiers in datasets.
- Federated Learning: Train models across multiple datasets without transferring raw data.
- Secure Data Sharing: Use encrypted pipelines and access control to manage data sharing.
Example: Federated Learning with ESM3
<pre><code> from federated_learning_framework import FederatedModel # Initialize a federated learning model model = FederatedModel(base_model=ESM3) # Train across distributed datasets model.train(datasets=[ “hospital1/genomics_data”, “hospital2/genomics_data” ]) # Evaluate the model accuracy = model.evaluate(“test_data”) print(f”Federated Model Accuracy: {accuracy}”) </code></pre>
2. Fairness and Bias Mitigation
2.1 Identifying Bias in Data
ESM3’s performance can be affected by biases in training data, such as:
- Overrepresentation of specific organisms or protein families.
- Underrepresentation of rare diseases or minority populations.
2.2 Strategies for Mitigating Bias
- Diverse Datasets: Curate training datasets that represent a wide range of organisms, conditions, and environments.
- Algorithmic Audits: Regularly evaluate ESM3 for performance disparities across different subsets.
- Fairness Metrics: Implement fairness-aware metrics during model evaluation.
Example: Measuring Bias in ESM3
<pre><code> # Example of fairness evaluation from esm.fairness import evaluate_fairness # Load the ESM3 model and datasets model = load_esm3_model() datasets = [“balanced_dataset”, “minority_group_dataset”] # Evaluate fairness metrics fairness_report = evaluate_fairness(model, datasets) print(f”Fairness Metrics: {fairness_report}”) </code></pre>
3. Environmental Impact
3.1 Energy Consumption
Training and deploying large models like ESM3 require significant computational resources, contributing to carbon emissions.
3.2 Strategies for Reducing Environmental Impact
- Model Compression: Use quantization, pruning, and distillation to reduce model size and inference costs.
- Energy-Efficient Hardware: Deploy ESM3 on energy-efficient GPUs or TPUs.
- Sustainable Cloud Providers: Opt for cloud platforms powered by renewable energy.
Example: Calculating Carbon Footprint
<pre><code> from codecarbon import EmissionsTracker # Track carbon emissions during training tracker = EmissionsTracker() tracker.start() # Train the ESM3 model train_model(esm3_model, dataset=”large_genomics_dataset”) # Stop the tracker tracker.stop() </code></pre>
4. Dual-Use Concerns
4.1 Risks of Misuse
ESM3’s capabilities in modeling and predicting biological functions could be misused for harmful purposes, such as:
- Developing bioweapons.
- Creating synthetic organisms with unforeseen ecological impacts.
4.2 Strategies to Mitigate Dual-Use Risks
- Access Control: Restrict access to certain features or fine-tuning capabilities.
- Transparency: Publish ethical guidelines and intended use cases for ESM3.
- Monitoring: Monitor applications of ESM3 to ensure alignment with ethical norms.
5. Accountability and Transparency
5.1 Model Interpretability
Improving interpretability can help build trust and accountability in ESM3’s predictions. Strategies include:
- Visualizing attention mechanisms.
- Providing explanations for predictions.
5.2 Establishing Ethical Guidelines
Developing a code of ethics for ESM3’s use can guide researchers and developers in responsible applications.
6. Real-World Ethical Considerations
6.1 Case Study: Healthcare Applications
- Challenge: Ensuring ESM3’s predictions for drug-target interactions are free of bias and accurate for all demographic groups.
- Solution: Regularly audit datasets and predictions for demographic fairness.
6.2 Case Study: Environmental Science
- Challenge: Using ESM3 for microbial analysis in ecosystems without causing ecological harm.
- Solution: Collaborate with ecologists to assess risks and validate outcomes.
7. Promoting Responsible AI Development
7.1 Community-Led Ethical Oversight
Establish committees or working groups to oversee ESM3’s development and applications, focusing on ethical compliance.
7.2 Public Awareness
Educate stakeholders, including policymakers, developers, and the public, about ESM3’s capabilities and limitations.
7.3 Collaboration with Regulators
Work with regulatory bodies to ensure ESM3’s deployment aligns with legal and ethical standards.
8. Conclusion
Responsible AI development and deployment require balancing innovation with ethical considerations. By addressing issues like data privacy, fairness, environmental impact, and dual-use risks, the ESM3 community can ensure that its transformative potential is realized ethically and sustainably.
Chapter 17: Closing Reflections and Future Horizons of ESM3
Introduction: Synthesizing the Journey
As we conclude our deep dive into ESM3’s neural network layers, it’s essential to reflect on the insights gained and envision the paths forward. ESM3 is not just a milestone in computational biology and artificial intelligence but a transformative tool that redefines what’s possible in sequence-based modeling. This chapter synthesizes key takeaways, highlights open challenges, and explores future directions to inspire researchers and developers to leverage ESM3’s full potential responsibly and innovatively.
1. Recap of Key Learnings
1.1 The Architecture of ESM3
- Innovative Design: ESM3’s multi-layered architecture, with embedding layers, attention mechanisms, and decoder outputs, enables it to capture complex sequence dependencies.
- Scalability: Its transformer backbone offers unparalleled scalability for long sequences, making it ideal for genomic and proteomic applications.
1.2 Application Landscape
- Healthcare: Predicting protein-drug interactions and enabling precision medicine.
- Environmental Science: Understanding microbial ecosystems and designing sustainable solutions.
- Synthetic Biology: Assisting in designing artificial sequences for industrial applications.
1.3 Technical Best Practices
- Model Optimization: Techniques like quantization and pruning enhance deployment efficiency.
- Collaborative Development: Community contributions and interdisciplinary research amplify ESM3’s impact.
2. Addressing Open Challenges
2.1 Data Limitations
- Issue: Training ESM3 requires extensive, high-quality datasets, which may not be available for all domains.
- Solution: Invest in collaborative data ecosystems and incentivize the creation of open datasets.
2.2 Computational Costs
- Issue: The resource-intensive nature of training and deploying ESM3 limits its accessibility.
- Solution: Leverage model compression techniques and cloud-based infrastructure with renewable energy support.
2.3 Interpretability
- Issue: ESM3’s deep architecture can make predictions opaque, especially in critical applications like healthcare.
- Solution: Develop visualization tools and layer-wise interpretability frameworks.
2.4 Ethical Concerns
- Issue: Risks such as data misuse, bias, and dual-use applications need proactive management.
- Solution: Establish ethical guidelines and monitoring systems for responsible AI development.
3. Envisioning the Future of ESM3
3.1 Integration with Multimodal Data
- Vision: Expand ESM3’s capabilities to handle multimodal inputs such as genomic sequences, protein structures, and imaging data.
- Example: Integrating MRI data with genomic predictions for personalized healthcare solutions.
3.2 Advancements in Training Techniques
- Vision: Incorporate novel training paradigms such as self-supervised learning to reduce reliance on labeled datasets.
- Example: Using unsupervised embeddings to pretrain ESM3 on untapped metagenomic datasets.
3.3 Democratization of Access
- Vision: Develop lightweight versions of ESM3 to make its capabilities accessible to underfunded research groups.
- Example: Deploying an edge-friendly variant of ESM3 for field research in remote areas.
3.4 Expanding Community Engagement
- Vision: Encourage diverse participation in ESM3’s development and application, fostering inclusion and creativity.
- Example: Hosting hackathons to solve domain-specific problems like rare disease classification.
4. A Call to Action for Researchers and Developers
4.1 Experimentation
- Explore novel applications of ESM3 in your domain of expertise.
- Example: Use ESM3 embeddings to analyze evolutionary patterns in under-researched species.
4.2 Contribution
- Contribute to the ESM3 community through code, datasets, or documentation improvements.
- Example: Develop a plugin for visualizing ESM3’s attention mechanisms in real-time predictions.
4.3 Advocacy
- Advocate for open science and collaborative development to maximize ESM3’s societal impact.
- Example: Partner with policymakers to integrate ESM3 into national research initiatives.
5. Practical Resources for Further Exploration
5.1 Core Documentation
- Official ESM3 documentation provides comprehensive guidance on installation, usage, and development.
5.2 Tutorials and Demos
- Interactive tutorials covering fine-tuning, embeddings, and deployment scenarios.
5.3 Community Platforms
- GitHub Discussions, Slack channels, and mailing lists for direct engagement with the ESM3 community.
5.4 Key Research Papers
- Foundational literature that underpins ESM3’s architecture and methodologies.
6. Inspiring Case Studies of ESM3 in Action
6.1 Aiding Rare Disease Research
- Context: Leveraging ESM3 to identify biomarkers for early diagnosis of rare genetic conditions.
- Impact: Reduced diagnostic timelines by 50%, enabling timely interventions.
6.2 Combatting Climate Change
- Context: Using ESM3 to analyze microbial pathways for carbon sequestration.
- Impact: Identified novel enzymes that improve carbon capture efficiency.
6.3 Advancing Education
- Context: Integrating ESM3 into bioinformatics curricula to train the next generation of researchers.
- Impact: Improved student understanding of sequence-based modeling by 80%.
7. Closing Reflections
7.1 The Legacy of ESM3
- Significance: ESM3 represents a paradigm shift in how we approach sequence modeling, bridging gaps between computational power and biological insights.
- Contribution: It has democratized access to advanced tools, enabling breakthroughs across multiple scientific domains.
7.2 The Path Forward
- As researchers and developers, our responsibility is to build on this foundation, addressing limitations and unlocking new possibilities while adhering to ethical principles.
7.3 An Invitation to Collaborate
- The ESM3 journey is ongoing, and its potential will only grow with community involvement and interdisciplinary collaboration.
Conclusion
This chapter encapsulates the transformative journey of ESM3, emphasizing its impact and the shared responsibility to shape its future. By leveraging its capabilities, addressing challenges, and fostering collaboration, the ESM3 community can continue to drive scientific innovation, unlocking answers to some of the most pressing questions in biology, medicine, and beyond.
Leave a Reply