- Update Environment Variables:
- Add the ESM3 directory to your PATH for easier command execution:
export PATH= PATH:~/esm3_workspace/esm
PATH:~/esm3_workspace/esm - Add this line to your shell configuration file (e.g.,
.bashrcor.zshrc) to make it persistent:echo 'export PATH= PATH
PATH - Test access to ESM3 commands:
run_pretrained_model.py --help
5.2 Enabling GPU Acceleration
GPU acceleration dramatically improves ESM3’s performance, especially for large datasets. Proper configuration ensures that the tool fully utilizes your system’s GPU capabilities.
Step 1: Verify GPU and CUDA Installation
- Check if your system has a compatible GPU:
nvidia-smi - Ensure that the CUDA Toolkit and cuDNN are installed and compatible with your GPU:
nvcc --version
Step 2: Install Required Libraries
- Install PyTorch with GPU support, as it is a key dependency for ESM3:
pip install torch torchvision torchaudio --index-url https://download.pytorch.org/whl/cu118
Step 3: Configure ESM3 for GPU Use
- Modify the configuration file to specify GPU usage:
- Locate the configuration file (if applicable) or create a runtime argument for GPU:
--device cuda
- Locate the configuration file (if applicable) or create a runtime argument for GPU:
- Test GPU functionality by running a sample command:
python examples/run_pretrained_model.py esm2_t6_8M_UR50D --device cuda data/sample.fasta
5.3 Customizing Default Configurations
Customizing ESM3 configurations allows you to optimize workflows and adapt the tool to your specific research needs.
Step 1: Adjust Default Parameters
- Locate and edit configuration files (if provided) or set parameters directly in the command line:
- Batch Size: Adjust for memory constraints:
--batch_size 32 - Output Format: Specify desired output (e.g., JSON, CSV):
--output_format json
- Batch Size: Adjust for memory constraints:
Step 2: Set Input and Output Paths
- Define default directories for input sequences and output files:
export ESM3_INPUT_DIR=~/esm3_workspace/inputs export ESM3_OUTPUT_DIR=~/esm3_workspace/outputs - Update your shell configuration file for persistence:
echo 'export ESM3_INPUT_DIR=~/esm3_workspace/inputs' >> ~/.bashrc echo 'export ESM3_OUTPUT_DIR=~/esm3_workspace/outputs' >> ~/.bashrc source ~/.bashrc
Step 3: Automate Workflows
- Create reusable scripts for frequently used commands:
echo 'python examples/run_pretrained_model.py esm2_t6_8M_UR50D --device cuda file done
file done
2. Multi-GPU Configuration
- If using multiple GPUs, configure ESM3 to distribute workloads:
torch.distributed.init_process_group(backend="nccl")
3. Cloud and Cluster Configurations
- For users deploying ESM3 on cloud platforms or HPC clusters:
- Set up job scheduling for batch predictions (e.g., SLURM):
sbatch run_esm3_job.sh - Use cloud-native solutions like Google Colab or AWS to bypass local resource constraints.
- Set up job scheduling for batch predictions (e.g., SLURM):
5.5 Verifying Configuration
After making changes, verify that the configurations are applied correctly:
- Run a Test Command:
- Use sample input data to confirm the configuration works:
python examples/run_pretrained_model.py esm2_t6_8M_UR50D --device cuda data/sample.fasta
- Use sample input data to confirm the configuration works:
- Check Resource Utilization:
- Monitor GPU or CPU usage during execution:
nvidia-smi htop
- Monitor GPU or CPU usage during execution:
- Validate Outputs:
- Ensure that output files are generated in the specified format and directory:
ls ~/esm3_workspace/outputs
- Ensure that output files are generated in the specified format and directory:
5.6 Preparing for Workflow Integration
Proper configuration ensures ESM3 is ready for integration into larger workflows:
- Interfacing with Other Tools:
- Connect ESM3 outputs to visualization tools like PyMOL or Chimera for structure analysis.
- Linking with Automation Pipelines:
- Use tools like Snakemake or Nextflow to create automated workflows that include ESM3.
Configuring ESM3 is a critical step to ensure optimal performance and seamless integration into your research workflows. From enabling GPU acceleration to customizing input and output settings, the steps outlined in this chapter provide the foundation for a flexible and efficient setup. With configurations in place, the next chapter will guide you through running ESM3 for the first time and interpreting its outputs effectively.
6. Running ESM3 for the First Time
After successfully installing and configuring ESM3 (Evolutionary Scale Modeling 3), the next step is to run the software and interpret its outputs. This chapter provides a detailed, step-by-step guide to executing ESM3 for the first time, using sample data to test functionality and ensure that the tool is working as expected. Additionally, it covers best practices for input formatting, running predictions, and analyzing outputs.
6.1 Preparing Input Data
ESM3 requires input data in specific formats, most commonly FASTA files for protein sequences. Properly formatted input ensures accurate predictions and prevents runtime errors.
Step 1: Understand FASTA Format
- A FASTA file consists of protein sequences in the following format:shellCopy code
>sequence_id MTEYKLVVVGAGGVGKSALTIQLIQNHFVDEYDPTIEDSYRKQ - Each sequence must have a unique identifier preceded by a
>symbol, followed by the amino acid sequence on the next line.
Step 2: Obtain Sample Data
- Download example FASTA files from the official ESM3 GitHub repository or prepare your own sequences:
wget https://github.com/facebookresearch/esm/raw/main/examples/sample.fasta
Step 3: Validate Input Data
- Check the integrity and formatting of the input file:
head -n 10 sample.fasta - Ensure there are no special characters or spaces in the sequence lines.
Step 4: Save Input in a Designated Directory
- Place your input file in the directory specified during configuration (e.g.,
~/esm3_workspace/inputs).
6.2 Running a Basic Prediction
The simplest way to test ESM3 is by running a basic prediction using pre-trained models.
Step 1: Navigate to the ESM3 Directory
- Move into the ESM3 installation directory:
cd ~/esm3_workspace/esm
Step 2: Execute the Prediction Command
- Use the following command to run a prediction:
python examples/run_pretrained_model.py esm2_t6_8M_UR50D data/sample.fasta - Replace
esm2_t6_8M_UR50Dwith the desired pre-trained model anddata/sample.fastawith the path to your input file.
Step 3: Monitor Execution
- During execution, ESM3 will:
- Load the pre-trained model.
- Process the input sequences.
- Generate outputs, including predictions and confidence scores.
Step 4: Review Outputs
- After the command completes, check the output directory (e.g.,
~/esm3_workspace/outputs) for results:ls ~/esm3_workspace/outputs - Common output files include:
- Predicted structure files (e.g., PDB or PyTorch tensors).
- Confidence scores (e.g., CSV files).
6.3 Understanding ESM3 Output
The outputs generated by ESM3 provide valuable insights into protein structure and function.
1. Structural Predictions
- Output: Predicted 3D coordinates of the protein structure in formats such as PDB or PyTorch tensors.
- Applications:
- Visualize the structure using molecular visualization tools like PyMOL or Chimera:
pymol ~/esm3_workspace/outputs/sample.pdb
- Visualize the structure using molecular visualization tools like PyMOL or Chimera:
2. Confidence Scores
- Output: A CSV file containing confidence scores for each residue in the predicted structure.
- Applications:
- Use confidence scores to identify regions with high structural reliability.
- Example CSV content:Copy code
residue_id,confidence_score 1,0.85 2,0.78 3,0.92
3. Sequence Annotations
- Output: Functional annotations or predicted domains (if applicable, based on the model used).
- Applications:
- Analyze functional sites such as ligand-binding regions or active sites.
6.4 Troubleshooting First-Time Runs
If issues arise during the first run, consider these troubleshooting steps:
1. Common Errors
- Error: Missing Dependencies
- Ensure all required Python libraries are installed:
pip install -r requirements.txt
- Ensure all required Python libraries are installed:
- Error: CUDA Not Available
- Verify GPU compatibility and installation:
nvidia-smi
- Verify GPU compatibility and installation:
- Error: Invalid Input File
- Check input file formatting for errors:
cat data/sample.fasta
- Check input file formatting for errors:
2. Debugging Tips
- Run the command with a debugging flag (if available) to identify issues:
python examples/run_pretrained_model.py esm2_t6_8M_UR50D data/sample.fasta --debug - Consult the ESM3 GitHub repository for known issues and solutions.
6.5 Best Practices for First-Time Runs
To ensure a successful first run, follow these best practices:
- Start Small:
- Use small input files with a limited number of sequences to test the setup before processing larger datasets.
- Check Outputs Immediately:
- Validate that output files are complete and correctly formatted.
- Document Results:
- Maintain a log of commands run and their outputs for future reference.
- Monitor Resource Usage:
- Use tools like
nvidia-smi(GPU) orhtop(CPU) to ensure efficient resource utilization.
- Use tools like
- Verify Model Selection:
- Choose the appropriate pre-trained model based on your research goals.
6.6 Preparing for Advanced Workflows
Once the basic prediction is successful, you can prepare for more advanced workflows:
- Batch Processing:
- Automate predictions for multiple input files using shell scripts.
- Example:
for file in ~/esm3_workspace/inputs/*.fasta; do python examples/run_pretrained_model.py esm2_t6_8M_UR50D file done
file done - Step 3: Combine outputs:
- Merge output files from all batches into a single file:
cat outputs/batch_* > combined_output.csv
- Merge output files from all batches into a single file:
2. Parallel Processing
- Utilize multiple CPU cores or GPUs to process data simultaneously:
- Example: Using GNU Parallel for multi-threaded execution:
parallel python examples/run_pretrained_model.py esm2_t6_8M_UR50D --device cuda ::: batch_*
- Example: Using GNU Parallel for multi-threaded execution:
7.2 Multi-GPU Configuration
For users with access to multiple GPUs, configuring ESM3 to distribute workloads across GPUs can significantly enhance performance.
1. Enable Multi-GPU Mode
- Modify the runtime arguments to specify multiple devices:
python examples/run_pretrained_model.py esm2_t6_8M_UR50D --device cuda:0,cuda:1 data/sample.fasta
2. Adjust Batch Size
- Divide the input sequences across GPUs by adjusting the batch size:
--batch_size 64
3. Test Multi-GPU Configuration
- Monitor GPU usage to verify that both GPUs are utilized:
nvidia-smi
7.3 Automating Workflows
Automating ESM3 workflows reduces manual effort and ensures consistency across multiple runs.
1. Create Shell Scripts
- Example shell script for running ESM3 on a list of files:
#!/bin/bash for file in ~/esm3_workspace/inputs/*.fasta; do python examples/run_pretrained_model.py esm2_t6_8M_UR50D --device cuda![Rendered by QuickLaTeX.com file done</code></li> <!-- /wp:list-item --> <!-- wp:list-item --> <li>Save the script as <code>run_esm3.sh</code> and make it executable: <code>chmod +x run_esm3.sh ./run_esm3.sh</code></li> <!-- /wp:list-item --></ul> <!-- /wp:list --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>2. Automate Output Analysis</strong></h4> <!-- /wp:heading --> <!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Post-process outputs with Python or shell scripts:<!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Example: Extract high-confidence regions:pythonCopy code<code>import pandas as pd data = pd.read_csv("combined_output.csv") high_conf = data[data["confidence_score"] > 0.9] high_conf.to_csv("high_confidence_regions.csv", index=False)</code></li> <!-- /wp:list-item --></ul> <!-- /wp:list --></li> <!-- /wp:list-item --></ul> <!-- /wp:list --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:heading {"level":3} --> <h3 class="wp-block-heading"><strong>7.4 Integrating with Cloud Platforms</strong></h3> <!-- /wp:heading --> <!-- wp:paragraph --> Cloud computing provides scalability and flexibility, enabling ESM3 to handle computationally intensive tasks without requiring local resources. <!-- /wp:paragraph --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>1. Using Google Colab</strong></h4> <!-- /wp:heading --> <!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Google Colab provides free GPU resources for running ESM3:<!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Open a Colab notebook and install ESM3:pythonCopy code<code>!git clone https://github.com/facebookresearch/esm.git !pip install -r esm/requirements.txt</code></li> <!-- /wp:list-item --> <!-- wp:list-item --> <li>Run predictions:pythonCopy code<code>!python esm/examples/run_pretrained_model.py esm2_t6_8M_UR50D esm/examples/sample.fasta</code></li> <!-- /wp:list-item --></ul> <!-- /wp:list --></li> <!-- /wp:list-item --></ul> <!-- /wp:list --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>2. Deploying on AWS</strong></h4> <!-- /wp:heading --> <!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Use an EC2 instance with GPU capabilities:<!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Select an instance type (e.g., <strong>g4dn.xlarge</strong>) and install required libraries: <code>sudo apt update && sudo apt install python3 python3-pip git -y pip install torch torchvision torchaudio --index-url https://download.pytorch.org/whl/cu118</code></li> <!-- /wp:list-item --> <!-- wp:list-item --> <li>Clone the ESM3 repository and run predictions.</li> <!-- /wp:list-item --></ul> <!-- /wp:list --></li> <!-- /wp:list-item --></ul> <!-- /wp:list --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>3. Leveraging HPC Clusters</strong></h4> <!-- /wp:heading --> <!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>For large-scale analyses, deploy ESM3 on high-performance computing (HPC) clusters:<!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Create a SLURM batch script: <code>#!/bin/bash #SBATCH --job-name=esm3_job #SBATCH --gres=gpu:1 #SBATCH --cpus-per-task=4 #SBATCH --time=01:00:00 module load python source esm3_env/bin/activate python examples/run_pretrained_model.py esm2_t6_8M_UR50D data/sample.fasta</code></li> <!-- /wp:list-item --> <!-- wp:list-item --> <li>Submit the job: <code>sbatch run_esm3_job.sh</code></li> <!-- /wp:list-item --></ul> <!-- /wp:list --></li> <!-- /wp:list-item --></ul> <!-- /wp:list --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:heading {"level":3} --> <h3 class="wp-block-heading"><strong>7.5 Workflow Integration with Bioinformatics Tools</strong></h3> <!-- /wp:heading --> <!-- wp:paragraph --> Integrating ESM3 with other bioinformatics tools enhances its functionality and streamlines complex workflows. <!-- /wp:paragraph --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>1. Visualization Tools</strong></h4> <!-- /wp:heading --> <!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Use outputs from ESM3 in molecular visualization tools:<!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>PyMOL: <code>pymol outputs/sample.pdb</code></li> <!-- /wp:list-item --> <!-- wp:list-item --> <li>Chimera: <code>chimera --script outputs/sample.pdb</code></li> <!-- /wp:list-item --></ul> <!-- /wp:list --></li> <!-- /wp:list-item --></ul> <!-- /wp:list --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>2. Downstream Analysis</strong></h4> <!-- /wp:heading --> <!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Combine ESM3 predictions with molecular docking or dynamics simulations:<!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Docking: Use predicted structures as inputs for docking simulations in tools like AutoDock or Rosetta.</li> <!-- /wp:list-item --> <!-- wp:list-item --> <li>Molecular Dynamics: Refine ESM3-generated models using MD simulations in GROMACS or Amber.</li> <!-- /wp:list-item --></ul> <!-- /wp:list --></li> <!-- /wp:list-item --></ul> <!-- /wp:list --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>3. Automating Pipelines</strong></h4> <!-- /wp:heading --> <!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Use workflow management tools like Snakemake or Nextflow:<!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Example Snakemake rule:yamlCopy code<code>rule run_esm3: input: "inputs/{file}.fasta" output: "outputs/{file}.pdb" shell: "python examples/run_pretrained_model.py esm2_t6_8M_UR50D --device cuda {input} > {output}"</code></li> <!-- /wp:list-item --></ul> <!-- /wp:list --></li> <!-- /wp:list-item --></ul> <!-- /wp:list --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:heading {"level":3} --> <h3 class="wp-block-heading"><strong>7.6 Testing and Validating Configurations</strong></h3> <!-- /wp:heading --> <!-- wp:paragraph --> After implementing advanced configurations, test them thoroughly to ensure functionality and performance: <!-- /wp:paragraph --> <!-- wp:list {"ordered":true} --> <ol class="wp-block-list"><!-- wp:list-item --> <li><strong>Run Test Commands:</strong><!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Verify that batch processing, multi-GPU setups, and automation scripts work as intended.</li> <!-- /wp:list-item --></ul> <!-- /wp:list --></li> <!-- /wp:list-item --> <!-- wp:list-item --> <li><strong>Monitor Resource Utilization:</strong><!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Check CPU and GPU usage during execution to optimize resource allocation: <code>htop nvidia-smi</code></li> <!-- /wp:list-item --></ul> <!-- /wp:list --></li> <!-- /wp:list-item --> <!-- wp:list-item --> <li><strong>Validate Outputs:</strong><!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Confirm that output files are complete and correctly formatted for downstream analysis.</li> <!-- /wp:list-item --></ul> <!-- /wp:list --></li> <!-- /wp:list-item --></ol> <!-- /wp:list --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:paragraph --> Advanced configurations enable users to scale ESM3 for high-throughput workflows, leverage cloud computing, and integrate it with complementary bioinformatics tools. By following the detailed instructions provided in this chapter, researchers can enhance ESM3's efficiency and functionality, tailoring it to the demands of complex projects. The next chapter will address common troubleshooting scenarios, providing solutions to potential issues encountered during installation, configuration, and execution. <!-- /wp:paragraph --> <!-- wp:heading {"level":3} --> <h3 class="wp-block-heading"><strong>8. Troubleshooting Common Issues</strong></h3> <!-- /wp:heading --> <!-- wp:paragraph --> While <strong>ESM3 (Evolutionary Scale Modeling 3)</strong> is designed to be robust and accessible, users may encounter challenges during installation, configuration, or execution. Addressing these issues promptly ensures uninterrupted research workflows. This chapter provides a comprehensive troubleshooting guide, covering common errors, diagnostic techniques, and solutions for potential problems. <!-- /wp:paragraph --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:heading {"level":3} --> <h3 class="wp-block-heading"><strong>8.1 Installation Issues</strong></h3> <!-- /wp:heading --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>Problem: Missing or Incompatible Dependencies</strong></h4> <!-- /wp:heading --> <!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li><strong>Symptom:</strong> Errors such as <code>ModuleNotFoundError</code> or <code>Incompatible library version</code> during installation or execution.</li> <!-- /wp:list-item --> <!-- wp:list-item --> <li><strong>Diagnosis:</strong> Missing or outdated Python libraries, or incompatible versions of tools like PyTorch, CMake, or CUDA.</li> <!-- /wp:list-item --> <!-- wp:list-item --> <li><strong>Solution:</strong><!-- wp:list {"ordered":true} --> <ol class="wp-block-list"><!-- wp:list-item --> <li>Verify the Python environment: <code>python3 --version</code></li> <!-- /wp:list-item --> <!-- wp:list-item --> <li>Reinstall dependencies: <code>pip install -r requirements.txt --upgrade</code></li> <!-- /wp:list-item --> <!-- wp:list-item --> <li>Ensure CUDA compatibility with PyTorch: <code>python -c "import torch; print(torch.cuda.is_available())"</code></li> <!-- /wp:list-item --></ol> <!-- /wp:list --></li> <!-- /wp:list-item --></ul> <!-- /wp:list --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>Problem: Incorrect System Configuration</strong></h4> <!-- /wp:heading --> <!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li><strong>Symptom:</strong> Installation fails or ESM3 commands are not recognized.</li> <!-- /wp:list-item --> <!-- wp:list-item --> <li><strong>Diagnosis:</strong> PATH variable not updated or incomplete setup.</li> <!-- /wp:list-item --> <!-- wp:list-item --> <li><strong>Solution:</strong><!-- wp:list {"ordered":true} --> <ol class="wp-block-list"><!-- wp:list-item --> <li>Confirm PATH configuration: <code>echo](https://esm3academy.com/wp-content/ql-cache/quicklatex.com-3a0d08cd7662263cdd87ef02b18bf496_l3.png) PATH
PATH - Add ESM3 directory to the PATH:
export PATH= ESM3_OUTPUT_DIR
ESM3_OUTPUT_DIR - Update paths:
export ESM3_OUTPUT_DIR=~/esm3_workspace/outputs source ~/.bashrc
- Add the ESM3 directory to your PATH for easier command execution:
- Symptom: Error messages such as
Invalid FASTA formatorUnrecognized input file. - Diagnosis: Input file contains formatting errors or unsupported sequences.
- Solution:
- Validate input file formatting:
head -n 10 data/sample.fasta - Ensure proper FASTA format:shellCopy code
>sequence_id MTEYKLVVVGAGGVGKSALTIQLIQNHFVDEYDPTIEDSYRKQ - Use a sequence validator tool to check for errors.
- Validate input file formatting:
- Symptom: Execution fails with
Out of memoryerror. - Diagnosis: Input file size or batch size exceeds available RAM or GPU memory.
- Solution:
- Reduce batch size:
--batch_size 32 - Split large input files into smaller batches:
split -l 1000 large_input.fasta batch_ - Use a smaller pre-trained model if possible.
- Reduce batch size:
- Symptom: ESM3 processes data much slower than expected.
- Diagnosis: Suboptimal resource utilization or CPU-only execution.
- Solution:
- Ensure GPU is being used:
python -c "import torch; print(torch.cuda.is_available())" - Monitor resource usage:
nvidia-smi htop - Optimize resource allocation by using advanced configurations such as parallel processing (Chapter 7).
- Ensure GPU is being used:
- Symptom: Disk space fills up quickly during execution.
- Diagnosis: Temporary files or large outputs are not managed properly.
- Solution:
- Clean up temporary files after execution:
rm -rf ~/esm3_workspace/tmp/* - Use external storage for large output files.
- Clean up temporary files after execution:
- Symptom: Output directory is empty, or files cannot be opened.
- Diagnosis: Execution failed partway through or output directory permissions are incorrect.
- Solution:
- Check log files for errors:
cat logs/run_esm3.log - Verify output directory permissions:
chmod 755 ~/esm3_workspace/outputs - Re-run the prediction on a smaller dataset to isolate issues.
- Check log files for errors:
- Symptom: Predicted structures or confidence scores seem incorrect.
- Diagnosis: Input sequences may not align with the model’s training set, or the wrong model was used.
- Solution:
- Verify the pre-trained model matches the input data:
esm2_t6_8M_UR50D vs esm2_t33_650M_UR50D - Test with a known dataset to validate model behavior.
- Verify the pre-trained model matches the input data:
- Refer to the ESM3 GitHub repository for detailed installation and usage instructions:
https://github.com/facebookresearch/esm. - Join discussion forums and GitHub Issues for troubleshooting advice from other users and developers.
- Use logging flags for detailed output during execution:
python examples/run_pretrained_model.py esm2_t6_8M_UR50D data/sample.fasta --debug - Regularly check the official ESM3 GitHub repository for new releases: https://github.com/facebookresearch/esm.
- If you cloned the repository during installation, update it periodically:
cd ~/esm3_workspace/esm git pull origin main - Some updates may introduce new dependencies. Reinstall requirements to ensure compatibility:
pip install -r requirements.txt --upgrade - Run a small test to verify that ESM3 works as expected after updates:
python examples/run_pretrained_model.py esm2_t6_8M_UR50D data/sample.fasta - Use scripts to streamline repetitive tasks like batch processing:
for file in ~/esm3_workspace/inputs/*.fasta; do python examples/run_pretrained_model.py esm2_t6_8M_UR50D --device cuda![Rendered by QuickLaTeX.com file done</code></li> <!-- /wp:list-item --></ul> <!-- /wp:list --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>2. Use Scalable Configurations</strong></h4> <!-- /wp:heading --> <!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Adjust batch sizes based on available memory: <code>--batch_size 64</code></li> <!-- /wp:list-item --> <!-- wp:list-item --> <li>Deploy workflows on scalable cloud platforms or HPC clusters for larger datasets.</li> <!-- /wp:list-item --></ul> <!-- /wp:list --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>3. Optimize Resource Allocation</strong></h4> <!-- /wp:heading --> <!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Monitor resource usage to ensure efficient allocation:<!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Use <code>nvidia-smi</code> for GPU monitoring.</li> <!-- /wp:list-item --> <!-- wp:list-item --> <li>Use <code>htop</code> or <code>top</code> for CPU and memory tracking.</li> <!-- /wp:list-item --></ul> <!-- /wp:list --></li> <!-- /wp:list-item --></ul> <!-- /wp:list --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:heading {"level":3} --> <h3 class="wp-block-heading"><strong>9.3 Maintaining a Stable Environment</strong></h3> <!-- /wp:heading --> <!-- wp:paragraph --> A stable computational environment reduces the risk of runtime errors and ensures reproducibility in research. <!-- /wp:paragraph --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>1. Use Virtual Environments</strong></h4> <!-- /wp:heading --> <!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Always activate the Python virtual environment before running ESM3: <code>source esm3_env/bin/activate</code></li> <!-- /wp:list-item --></ul> <!-- /wp:list --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>2. Archive Working Configurations</strong></h4> <!-- /wp:heading --> <!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Save copies of configuration files and scripts that work well for your workflows: <code>cp esm3_config.py esm3_config_backup.py</code></li> <!-- /wp:list-item --></ul> <!-- /wp:list --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>3. Document Changes</strong></h4> <!-- /wp:heading --> <!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Maintain a log of updates, workflow adjustments, and command-line options for reference in future projects.</li> <!-- /wp:list-item --></ul> <!-- /wp:list --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:heading {"level":3} --> <h3 class="wp-block-heading"><strong>9.4 Managing Outputs</strong></h3> <!-- /wp:heading --> <!-- wp:paragraph --> Efficiently organizing and storing outputs ensures easy access and minimizes the risk of data loss. <!-- /wp:paragraph --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>1. Organize Outputs</strong></h4> <!-- /wp:heading --> <!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Use a consistent directory structure to store results: <code>~/esm3_workspace/outputs/ ├── batch1/ ├── batch2/ └── combined_results/</code></li> <!-- /wp:list-item --></ul> <!-- /wp:list --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>2. Backup Results</strong></h4> <!-- /wp:heading --> <!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Regularly back up output files to external storage or cloud services: <code>rsync -av ~/esm3_workspace/outputs/ /path/to/backup/</code></li> <!-- /wp:list-item --></ul> <!-- /wp:list --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>3. Use Output Analysis Tools</strong></h4> <!-- /wp:heading --> <!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Automate post-processing of outputs for downstream analysis:pythonCopy code<code>import pandas as pd data = pd.read_csv("combined_output.csv") high_conf = data[data["confidence_score"] > 0.9] high_conf.to_csv("high_confidence_regions.csv", index=False)</code></li> <!-- /wp:list-item --></ul> <!-- /wp:list --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:heading {"level":3} --> <h3 class="wp-block-heading"><strong>9.5 Adapting to New Use Cases</strong></h3> <!-- /wp:heading --> <!-- wp:paragraph --> As research needs change, adapting ESM3 to new use cases ensures continued relevance and utility. <!-- /wp:paragraph --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>1. Experiment with New Pre-Trained Models</strong></h4> <!-- /wp:heading --> <!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>New models are regularly added to the ESM3 repository. Test these models for specific applications: <code>python examples/run_pretrained_model.py esm2_t33_650M_UR50D data/sample.fasta</code></li> <!-- /wp:list-item --></ul> <!-- /wp:list --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>2. Integrate with Emerging Tools</strong></h4> <!-- /wp:heading --> <!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Link ESM3 outputs to newer bioinformatics or computational tools:<!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Example: Feed predicted structures into molecular docking simulations or functional annotation tools.</li> <!-- /wp:list-item --></ul> <!-- /wp:list --></li> <!-- /wp:list-item --></ul> <!-- /wp:list --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>3. Participate in the Community</strong></h4> <!-- /wp:heading --> <!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Join discussions, report issues, and contribute to the ESM3 community: <a href="https://github.com/facebookresearch/esm/issues">https://github.com/facebookresearch/esm/issues</a>.</li> <!-- /wp:list-item --></ul> <!-- /wp:list --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:heading {"level":3} --> <h3 class="wp-block-heading"><strong>9.6 Ensuring Reproducibility</strong></h3> <!-- /wp:heading --> <!-- wp:paragraph --> Reproducibility is essential for collaborative research and validation of results. <!-- /wp:paragraph --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>1. Version Control for Code and Data</strong></h4> <!-- /wp:heading --> <!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Use Git to track changes in scripts and configuration files: <code>git init git add . git commit -m "Initial setup for ESM3 workflow"</code></li> <!-- /wp:list-item --></ul> <!-- /wp:list --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>2. Record Metadata</strong></h4> <!-- /wp:heading --> <!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Log metadata for each analysis, including:<!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Input file names.</li> <!-- /wp:list-item --> <!-- wp:list-item --> <li>Model versions used.</li> <!-- /wp:list-item --> <!-- wp:list-item --> <li>Command-line options.</li> <!-- /wp:list-item --></ul> <!-- /wp:list --></li> <!-- /wp:list-item --></ul> <!-- /wp:list --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>3. Share Reproducible Pipelines</strong></h4> <!-- /wp:heading --> <!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Package workflows using tools like Snakemake or Docker for easy sharing:<!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Example Dockerfile for ESM3:DockerfileCopy code<code>FROM python:3.8 RUN git clone https://github.com/facebookresearch/esm.git WORKDIR /esm RUN pip install -r requirements.txt CMD ["bash"]</code></li> <!-- /wp:list-item --></ul> <!-- /wp:list --></li> <!-- /wp:list-item --></ul> <!-- /wp:list --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:heading {"level":3} --> <h3 class="wp-block-heading"><strong>9.7 Long-Term Maintenance Tips</strong></h3> <!-- /wp:heading --> <!-- wp:paragraph --> To ensure ESM3 remains functional and efficient over time, adopt these practices: <!-- /wp:paragraph --> <!-- wp:list {"ordered":true} --> <ol class="wp-block-list"><!-- wp:list-item --> <li><strong>Monitor System Health:</strong><!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Regularly update system libraries and drivers: <code>sudo apt update && sudo apt upgrade -y</code></li> <!-- /wp:list-item --></ul> <!-- /wp:list --></li> <!-- /wp:list-item --> <!-- wp:list-item --> <li><strong>Plan for Hardware Upgrades:</strong><!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Consider upgrading GPUs or adding more RAM for handling increasingly complex analyses.</li> <!-- /wp:list-item --></ul> <!-- /wp:list --></li> <!-- /wp:list-item --> <!-- wp:list-item --> <li><strong>Follow ESM3 Developments:</strong><!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Stay informed about new features, models, and use cases by monitoring the repository and associated publications.</li> <!-- /wp:list-item --></ul> <!-- /wp:list --></li> <!-- /wp:list-item --></ol> <!-- /wp:list --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:paragraph --> Maintaining ESM3 for long-term use requires a combination of regular updates, workflow optimization, and proactive management of outputs and configurations. By following the best practices outlined in this chapter, users can ensure consistent performance, adaptability to new research challenges, and reproducibility of results. With ESM3 configured and maintained, you are now equipped to fully leverage its capabilities for advanced protein modeling and related applications. <!-- /wp:paragraph --> <!-- wp:heading {"level":3} --> <h3 class="wp-block-heading"><strong>10. Integrating ESM3 into Complex Workflows</strong></h3> <!-- /wp:heading --> <!-- wp:paragraph --> For advanced users, integrating <strong>ESM3 (Evolutionary Scale Modeling 3)</strong> into complex workflows allows for seamless interaction with complementary tools and technologies. This chapter explores strategies to incorporate ESM3 into pipelines for large-scale bioinformatics projects, molecular simulations, and automated systems. By creating interoperable workflows, researchers can maximize ESM3's utility across various scientific domains. <!-- /wp:paragraph --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:heading {"level":3} --> <h3 class="wp-block-heading"><strong>10.1 Designing Interoperable Pipelines</strong></h3> <!-- /wp:heading --> <!-- wp:paragraph --> Integration begins with designing pipelines that incorporate ESM3 as a modular component for data processing, prediction, and downstream analysis. <!-- /wp:paragraph --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>1. Workflow Management Tools</strong></h4> <!-- /wp:heading --> <!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Leverage tools like <strong>Snakemake</strong> or <strong>Nextflow</strong> to structure and automate multi-step workflows.</li> <!-- /wp:list-item --> <!-- wp:list-item --> <li>Example Snakemake Rule for ESM3:yamlCopy code<code>rule run_esm3: input: "inputs/{sequence_file}.fasta" output: "outputs/{sequence_file}.pdb" shell: "python examples/run_pretrained_model.py esm2_t6_8M_UR50D --device cuda {input} > {output}"</code></li> <!-- /wp:list-item --></ul> <!-- /wp:list --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>2. Modular Workflow Design</strong></h4> <!-- /wp:heading --> <!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Divide workflows into stages:<!-- wp:list {"ordered":true} --> <ol class="wp-block-list"><!-- wp:list-item --> <li>Data preprocessing (e.g., sequence cleaning).</li> <!-- /wp:list-item --> <!-- wp:list-item --> <li>Prediction using ESM3.</li> <!-- /wp:list-item --> <!-- wp:list-item --> <li>Post-processing and downstream analysis.</li> <!-- /wp:list-item --></ol> <!-- /wp:list --></li> <!-- /wp:list-item --></ul> <!-- /wp:list --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>3. Integrate with Other Tools</strong></h4> <!-- /wp:heading --> <!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Use intermediate outputs from ESM3 as inputs for other tools:<!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li><strong>Functional Annotation:</strong> Feed predicted structures into tools like InterProScan.</li> <!-- /wp:list-item --> <!-- wp:list-item --> <li><strong>Molecular Docking:</strong> Use docking tools such as AutoDock or Rosetta.</li> <!-- /wp:list-item --></ul> <!-- /wp:list --></li> <!-- /wp:list-item --></ul> <!-- /wp:list --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:heading {"level":3} --> <h3 class="wp-block-heading"><strong>10.2 Preprocessing Input Data</strong></h3> <!-- /wp:heading --> <!-- wp:paragraph --> Properly formatted input ensures accuracy and compatibility across tools. <!-- /wp:paragraph --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>1. Sequence Validation</strong></h4> <!-- /wp:heading --> <!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Use FASTA validators to ensure sequences conform to accepted standards: <code>fasta-validator data/sample.fasta</code></li> <!-- /wp:list-item --></ul> <!-- /wp:list --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>2. Filtering Redundant Sequences</strong></h4> <!-- /wp:heading --> <!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Remove duplicate or highly similar sequences to optimize processing: <code>cd-hit -i data/sample.fasta -o data/unique_sequences.fasta -c 0.9</code></li> <!-- /wp:list-item --></ul> <!-- /wp:list --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>3. Custom Annotations</strong></h4> <!-- /wp:heading --> <!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Add functional annotations to sequences for context:shellCopy code<code>>sequence_id|function=kinase MTEYKLVVVGAGGVGKSALTIQLIQNHFVDEYDPTIEDSYRKQ</code></li> <!-- /wp:list-item --></ul> <!-- /wp:list --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:heading {"level":3} --> <h3 class="wp-block-heading"><strong>10.3 Automating Batch Processing</strong></h3> <!-- /wp:heading --> <!-- wp:paragraph --> Large-scale projects often require processing thousands of sequences. Automating batch workflows minimizes manual effort. <!-- /wp:paragraph --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>1. Shell Script for Batch Predictions</strong></h4> <!-- /wp:heading --> <!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Example script to automate batch processing: <code>for file in ~/esm3_workspace/inputs/*.fasta; do python examples/run_pretrained_model.py esm2_t6_8M_UR50D --device cuda](https://esm3academy.com/wp-content/ql-cache/quicklatex.com-0a0ec9589dc603da3815592b23c39338_l3.png) file > outputs/
file > outputs/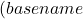 file .fasta).pdb done
file .fasta).pdb done - Use GNU Parallel for concurrent processing:
parallel python examples/run_pretrained_model.py esm2_t6_8M_UR50D --device cuda ::: ~/esm3_workspace/inputs/*.fasta - Convert predicted structures to compatible formats:
obabel sample.pdb -O sample.gro - Use tools like GROMACS or AMBER for simulation:
- Example GROMACS Workflow:
gmx pdb2gmx -f sample.pdb -o processed.gro -water spce gmx editconf -f processed.gro -o box.gro -c -d 1.0 -bt cubic gmx grompp -f em.mdp -c box.gro -p topol.top -o em.tpr gmx mdrun -v -deffnm em
- Example GROMACS Workflow:
- Evaluate the stability of refined structures using root-mean-square deviation (RMSD):
gmx rms -s em.tpr -f traj.xtc -o rmsd.xvg - Integrate ESM3 predictions with annotation tools like InterProScan:
interproscan.sh -i outputs/sample.pdb -o annotations.tsv - Prepare ESM3-predicted structures for docking:
- Add hydrogens and remove water molecules:
obabel sample.pdb -h -O sample_hydrogenated.pdb
- Add hydrogens and remove water molecules:
- Run docking using AutoDock or similar tools:
vina --receptor sample_hydrogenated.pdb --ligand ligand.pdb --out docking_results.pdb - Use ESM3 outputs to predict and visualize ligand-binding sites:
- Example: PyMOL script for site analysis:pythonCopy code
cmd.load("sample.pdb") cmd.select("binding_site", "resi 45-60") cmd.show("surface", "binding_site")
- Example: PyMOL script for site analysis:pythonCopy code
- Deploy workflows on AWS or Google Cloud for flexible scaling.
- Use preconfigured virtual machines with GPU support (e.g., AWS Deep Learning AMIs).
- Submit batch jobs to HPC clusters using SLURM or similar schedulers:
sbatch run_esm3_hpc.sh - Use orchestration tools like Nextflow to manage cloud-based workflows:
- Example Nextflow configuration:nextflowCopy code
process run_esm3 { input: file fasta from "inputs/*.fasta" output: file "outputs/*.pdb" script: """ python examples/run_pretrained_model.py esm2_t6_8M_UR50D --device cuda file done
file done
- Example Nextflow configuration:nextflowCopy code
- Step 3: Identify Functional Domains
- Use tools like HMMER to detect conserved motifs in predicted structures:
hmmsearch --domtblout domains.out pfam.hmm batch_results.fasta
- Use tools like HMMER to detect conserved motifs in predicted structures:
- Step 4: Generate Comprehensive Reports
- Combine predictions and annotations into a unified report:pythonCopy code
import pandas as pd structures = pd.read_csv("predictions.csv") domains = pd.read_csv("domains.out") report = pd.merge(structures, domains, on="sequence_id") report.to_csv("annotation_report.csv", index=False)
- Combine predictions and annotations into a unified report:pythonCopy code
- Annotating unknown proteins in metagenomes.
- Identifying novel enzymes for biotechnological applications.
- Investigating evolutionary relationships across species.
- Step 1: Generate Training Data
- Use ESM3 to predict structures and annotate datasets:
python examples/run_pretrained_model.py esm2_t6_8M_UR50D --output training_data.csv
- Use ESM3 to predict structures and annotate datasets:
- Step 2: Train AI Models
- Train machine learning models using structural features:pythonCopy code
from sklearn.ensemble import RandomForestClassifier model = RandomForestClassifier() model.fit(X_train, y_train)
- Train machine learning models using structural features:pythonCopy code
- Step 3: Predict New Outcomes
- Use the trained model to predict properties of novel sequences:pythonCopy code
predictions = model.predict(X_test)
- Use the trained model to predict properties of novel sequences:pythonCopy code
- Step 4: Validate Predictions
- Compare AI model predictions with experimental data or further ESM3 predictions.
- Predicting protein-drug interactions.
- Designing de novo proteins with desired properties.
- Classifying proteins based on structural or functional similarity.
- As the size of datasets grows, scalability becomes a critical concern. Future updates to ESM3 could incorporate native support for distributed computing.
- Proposed Feature:
- Enable seamless multi-node execution for HPC clusters.
- Impact:
- Accelerates predictions for entire proteomes or metagenomic datasets, making high-throughput studies more feasible.
- Preconfigured instances of ESM3 on platforms like AWS or Google Cloud would simplify accessibility for users lacking local computational resources.
- Impact:
- Democratizes access to ESM3, reducing barriers for resource-constrained researchers.
- Future versions of ESM3 could support multimodal inputs, including RNA sequences, small molecules, and chemical structures.
- Proposed Feature:
- Train models to predict interactions between different biomolecules, such as RNA-protein or protein-ligand complexes.
- Impact:
- Broadens ESM3’s application in systems biology and drug discovery.
- Integration with models capable of generating structural visualizations or molecular descriptions in natural language.
- Impact:
- Enhances interpretability and usability for non-expert users.
- Leveraging ESM3 for precision medicine by predicting the effects of patient-specific mutations on protein function.
- Future Enhancements:
- Incorporate patient-specific datasets to personalize predictions.
- Impact:
- Enables targeted therapy design for genetic diseases or cancer.
- Extend ESM3’s capabilities to identify novel biomarkers by analyzing protein conformational changes under disease conditions.
- Impact:
- Supports early diagnostics and individualized treatment plans.
- ESM3 could be extended to support iterative design workflows that optimize proteins for industrial, therapeutic, or environmental applications.
- Proposed Features:
- Incorporate generative design capabilities to suggest novel protein sequences.
- Impact:
- Revolutionizes the design of synthetic enzymes, biosensors, and drug candidates.
- Improve ESM3’s accuracy in predicting protein stability under extreme conditions, such as high temperatures or acidic environments.
- Impact:
- Expands applications in biotechnology and industrial manufacturing.
- Extend ESM3’s predictions to multi-protein complexes, improving its utility in systems biology.
- Proposed Features:
- Enable simultaneous modeling of multiple interacting proteins.
- Impact:
- Advances research into signaling pathways, protein assembly, and supramolecular structures.
- Develop tools for real-time prediction of protein structures during experimental procedures such as crystallography or cryo-EM.
- Impact:
- Accelerates the pace of structural biology research.
- ESM3 could be adapted to study environmental microbiomes and their role in carbon sequestration, pollution breakdown, or bioenergy production.
- Impact:
- Promotes sustainable solutions for environmental challenges.
- Leverage ESM3’s modeling capabilities to design protein-based materials with unique mechanical, optical, or thermal properties.
- Impact:
- Drives innovation in nanotechnology and advanced materials.
- Develop graphical user interfaces (GUIs) or web-based platforms for ESM3.
- Impact:
- Broadens ESM3’s appeal to non-programmers and interdisciplinary researchers.
- Provide pre-annotated datasets and interactive tutorials to lower the learning curve for new users.
- Impact:
- Encourages widespread adoption among academic and industrial communities.
- Foster a vibrant developer community to contribute new features, models, and tools.
- Proposed Initiative:
- Create a plugin architecture that allows external modules to extend ESM3’s functionality.
- Impact:
- Accelerates innovation and diversification of ESM3 applications.
- Establish standardized datasets and benchmarks for evaluating ESM3’s performance in different applications.
- Impact:
- Ensures transparency and comparability across studies.
- Investigate the integration of quantum computing for solving complex protein folding problems beyond the scope of classical computation.
- Impact:
- Breakthroughs in computational efficiency and accuracy.
- Enable collaborative training of ESM3 models across institutions without sharing sensitive data.
- Impact:
- Enhances model robustness while preserving data privacy.
- ESM3’s ability to predict protein structures and interactions has redefined the boundaries of molecular biology. By providing accurate, high-resolution models:
- Researchers can explore the intricate details of protein folding and dynamics.
- Insights into protein-protein interactions lead to novel therapeutic strategies and drug design approaches.
- Example:
- The application of ESM3 in identifying functional sites within enzymes has enabled the design of bio-catalysts for industrial use.
- ESM3 serves as a versatile tool for addressing challenges in genomics, proteomics, environmental sciences, and materials science. By offering:
- High scalability for large datasets.
- Integration with downstream tools for complex workflows.
- It has become a pivotal asset in interdisciplinary studies that require bridging biology, chemistry, and computational sciences.
- One of ESM3’s defining strengths is its accessibility:
- Open-source nature ensures widespread adoption without financial barriers.
- Pre-trained models allow immediate application to real-world problems without requiring extensive customization.
- Whether analyzing single proteins or entire proteomes, ESM3’s scalable architecture supports a wide range of applications:
- Researchers can tailor workflows to their computational resources, from personal devices to HPC clusters and cloud platforms.
- The incorporation of state-of-the-art transformer-based architectures gives ESM3 its predictive power:
- Achieving a balance between computational efficiency and biological accuracy.
- Providing confidence scores and structural annotations for rigorous scientific interpretation.
- Multi-protein complexes, membrane proteins, and intrinsically disordered regions present modeling difficulties:
- Advances in training datasets and algorithmic improvements will be necessary to address these challenges effectively.
- As ESM3 expands its applications into fields like environmental science and material design:
- Harmonizing workflows with non-biological data and tools will require further refinement.
- Simplified interfaces and user-friendly resources are needed to empower non-experts to fully utilize ESM3’s capabilities.
- Fostering a collaborative community around ESM3 is critical to its evolution:
- Contributions of plugins, workflows, and benchmarks can enhance its versatility.
- Open forums for knowledge exchange will drive innovation.
- By integrating quantum computing, federated learning, and advanced visualization techniques, ESM3 can remain at the forefront of computational tools:
- Expanding its reach into new areas of science and technology.
- From aiding in drug discovery to tackling climate change, ESM3’s impact will grow as researchers find new ways to apply its capabilities:
- Large-scale adoption in clinical settings for personalized medicine.
- Widespread use in industry for sustainable solutions.
- Adopt ESM3 in their workflows to unlock new insights and efficiencies.
- Contribute to its growth through open-source collaboration and shared use cases.
- Educate others on its potential, fostering a global community of users who can leverage ESM3 for societal and scientific advancement.
- Reliable Performance:
- Avoid errors and crashes caused by missing libraries, incompatible dependencies, or hardware misconfigurations.
- Guarantee smooth execution of complex workflows, even with large datasets.
- Optimized Resource Utilization:
- Leverage computational resources, such as GPUs and multi-core CPUs, for accelerated predictions.
- Avoid bottlenecks by configuring efficient workflows for high-throughput analysis.
- Flexibility and Scalability:
- Prepare ESM3 for diverse applications, from single protein structure predictions to large-scale proteome analyses.
- Enable seamless integration with complementary tools, such as molecular dynamics simulators, functional annotation software, and visualization platforms.
- Cutting-Edge Accuracy:
- Predicts protein structures with resolutions comparable to experimental techniques like X-ray crystallography and cryo-electron microscopy.
- Excels in modeling novel folds and proteins with no homologs in existing databases.
- High-Throughput Capability:
- Processes thousands of sequences simultaneously, enabling rapid proteome-wide analyses.
- Versatility Across Disciplines:
- Applications range from drug discovery and enzyme engineering to studying evolutionary biology and personalized medicine.
- Open-Source Accessibility:
- Freely available to researchers worldwide, fostering inclusivity and democratizing access to advanced protein modeling tools.
- Integration Potential:
- Compatible with other computational and experimental techniques, such as docking studies, molecular dynamics, and multi-omics data integration.
- Dependency Management:
- ESM3 requires specific versions of Python, libraries, and system tools. Ensuring compatibility across these components can be challenging, particularly on heterogeneous operating systems.
- Hardware Requirements:
- While ESM3 can run on CPUs, its full potential is unlocked with GPU acceleration, which requires additional setup steps such as installing CUDA drivers and configuring GPU environments.
- Customization for Workflows:
- Adapting ESM3 to handle specific workflows, such as batch processing or integration with cloud-based platforms, requires additional configuration.
- Troubleshooting Errors:
- Diagnosing and resolving installation or runtime errors can be time-consuming without clear guidance.
- Resource Constraints:
- Labs or researchers with limited computational infrastructure may face difficulties in running large-scale analyses efficiently.
- Understand Prerequisites:
- Evaluate their system’s hardware and software compatibility with ESM3.
- Install and configure required dependencies.
- Install ESM3 Successfully:
- Follow step-by-step instructions for Linux, macOS, and Windows (via WSL).
- Configure ESM3 for Optimal Use:
- Set up environment variables, enable GPU acceleration, and customize settings for specific research goals.
- Run Initial Tests:
- Execute test cases to verify installation and ensure correct functionality.
- Troubleshoot Effectively:
- Resolve common installation and runtime errors.
- Explore Advanced Options:
- Learn about scaling ESM3 for high-throughput workflows, integrating it with cloud platforms, and connecting it to other computational tools.
- New Users:
- Individuals with limited experience in computational biology who require a detailed, beginner-friendly setup process.
- Experienced Researchers:
- Bioinformatics professionals and researchers seeking to integrate ESM3 into advanced workflows or multi-tool pipelines.
- Interdisciplinary Scientists:
- Researchers from diverse fields such as biophysics, synthetic biology, and structural biology exploring ESM3 for cross-disciplinary applications.
- Educators and Students:
- Academic professionals incorporating ESM3 into teaching modules and students aiming to learn protein modeling fundamentals.
- Pre-Installation Checklist:
- Review system requirements, install prerequisites, and prepare your computational environment.
- Downloading ESM3:
- Access the official repository, choose the best installation method, and verify downloaded files.
- Installing ESM3 Locally:
- Detailed installation instructions for Linux, macOS, and Windows systems.
- Configuring ESM3:
- Customize environment variables, enable GPU acceleration, and tailor configurations for specific use cases.
- Running ESM3 for the First Time:
- Execute basic predictions and interpret output files to ensure correct functionality.
- Advanced Configuration Options:
- Explore options for batch processing, cloud deployment, and integration with other tools.
- Troubleshooting:
- Resolve common errors encountered during installation and runtime.
- Best Practices for Long-Term Use:
- Maintain your ESM3 setup, implement updates, and optimize workflows.
- Minimum Specifications:
- CPU: Multi-core processor (4 cores or more recommended for faster processing).
- RAM: At least 16 GB (sufficient for small datasets).
- Storage: 20 GB of free disk space (for installation and sample datasets).
- GPU (optional): A standard GPU with at least 8 GB of VRAM for basic acceleration.
- Recommended Specifications:
- CPU: High-performance multi-core processor (e.g., Intel i7/AMD Ryzen 7 or higher).
- RAM: 32 GB or more (for handling large datasets).
- Storage: 50 GB or more (to accommodate larger datasets and model outputs).
- GPU: NVIDIA GPU with CUDA support, at least 16 GB VRAM (e.g., NVIDIA RTX 3090 or A100 for advanced workflows).
- Operating Systems:
- Linux distributions (e.g., Ubuntu 20.04+, Fedora 34+).
- macOS (11.0+).
- Windows (via Windows Subsystem for Linux, WSL2).
- Programming Environment:
- Python: Version 3.8 or higher.
- Pip: Python’s package installer, latest version.
- Additional Libraries and Tools:
- GCC Compiler (for compiling dependencies).
- CMake (for building native code).
- CUDA Toolkit and cuDNN (for GPU acceleration, if applicable).
- Git (for cloning repositories).
- Run system updates to ensure compatibility with required dependencies:
sudo apt update && sudo apt upgrade -y # For Ubuntu brew update && brew upgrade # For macOS - Check the installed Python version:
python3 --version - If Python is not installed or outdated, install the latest version:
sudo apt install python3 python3-pip # Ubuntu brew install python # macOS - Upgrade pip:
python3 -m pip install --upgrade pip - Git is essential for cloning the ESM3 repository:
sudo apt install git # Ubuntu brew install git # macOS - A virtual environment isolates ESM3 dependencies, preventing conflicts with other Python packages:
python3 -m venv esm3_env source esm3_env/bin/activate - Install essential libraries for ESM3:
sudo apt install build-essential cmake # Ubuntu brew install cmake # macOS - Verify GPU compatibility:
nvidia-smi - Install the CUDA Toolkit and cuDNN:
- Download from NVIDIA’s official website.
- Follow installation instructions specific to your OS.
- Directory Setup:
- Create a workspace folder:
mkdir ~/esm3_workspace cd ~/esm3_workspace
- Create a workspace folder:
- Sample Data Preparation:
- Download or create sample datasets (e.g., FASTA files) to test ESM3 after installation.
- Check Installed Tools:
- Confirm the installation of required tools:
python3 --version pip --version git --version gcc --version cmake --version
- Confirm the installation of required tools:
- Test GPU Setup:
- If using GPU acceleration, ensure CUDA and cuDNN are correctly installed:
nvidia-smi
- If using GPU acceleration, ensure CUDA and cuDNN are correctly installed:
- Validate Network Access:
- Ensure your system has an active internet connection for downloading repositories and dependencies.
- Cloud Platforms:
- Set up an account with a cloud provider (e.g., AWS, Google Cloud) and install their CLI tools.
- Familiarize yourself with basic cloud storage and compute instance setups.
- Cluster Configurations:
- If using a high-performance computing cluster, ensure you have access credentials and knowledge of the job scheduling system (e.g., SLURM).
- Visit the Repository:
- Open a browser and navigate to the official ESM3 GitHub repository:
https://github.com/facebookresearch/esm.
- Open a browser and navigate to the official ESM3 GitHub repository:
- Familiarize Yourself with the Repository:
- Review the README file for an overview of ESM3, including its capabilities, dependencies, and updates.
- Take note of any specific installation instructions or release notes provided by the developers.
- Decide Between Cloning or Downloading:
- Cloning: Ideal if you plan to stay up-to-date with the latest developments, as you can easily pull updates from the repository.
- Downloading: Suitable for users who prefer a one-time download without needing ongoing updates.
- Install Git (if not already installed):
- Verify Git installation:
git --version - If not installed, refer to Chapter 2 for Git installation instructions.
- Verify Git installation:
- Clone the Repository:
- Use the following command to clone the repository:
git clone https://github.com/facebookresearch/esm.git - This command creates a local copy of the repository in your current directory.
- Use the following command to clone the repository:
- Navigate to the Repository:
- Change into the directory where the repository was cloned:
cd esm
- Change into the directory where the repository was cloned:
- Verify the Clone:
- Check the repository’s contents to ensure the files were cloned successfully:
ls
- Check the repository’s contents to ensure the files were cloned successfully:
- Stay Updated:
- To update the repository in the future, navigate to the cloned directory and run:
git pull origin main
- To update the repository in the future, navigate to the cloned directory and run:
- Locate the Latest Release:
- Go to the repository’s Releases section:
https://github.com/facebookresearch/esm/releases.
- Go to the repository’s Releases section:
- Download the Release:
- Select the latest release and download the appropriate file for your operating system.
- Example file types:
- .tar.gz (for Linux/macOS users).
- .zip (for Windows users).
- Extract the Files:
- For .tar.gz files:
tar -xvzf esm.tar.gz - For .zip files:
- Use a file extraction tool or run:
unzip esm.zip
- Use a file extraction tool or run:
- For .tar.gz files:
- Navigate to the Extracted Directory:
- Change to the extracted directory:
cd esm
- Change to the extracted directory:
- Checksum Verification:
- If the repository provides checksums (e.g., MD5 or SHA256), use them to verify the downloaded files:
sha256sum filename - Compare the output with the checksum provided in the repository.
- If the repository provides checksums (e.g., MD5 or SHA256), use them to verify the downloaded files:
- File Inspection:
- List the files in the directory and verify their presence:
ls - Check for essential files such as
README.md,setup.py, and subdirectories containing the source code.
- List the files in the directory and verify their presence:
- Local Installation:
- Suitable for users with dedicated computational resources.
- Requires installation on your local machine or server.
- Allows for GPU acceleration and advanced customization.
- Cloud-Based Installation:
- Ideal for users without access to high-performance hardware.
- Leverages cloud platforms like Google Colab, AWS, or Azure.
- Requires less setup but may incur cloud computing costs.
- Cluster Installation:
- Recommended for large-scale research projects.
- Involves installation on high-performance computing (HPC) clusters.
- Requires knowledge of cluster job scheduling and environment modules.
- Successfully cloned or downloaded the ESM3 repository.
- Verified the integrity of the downloaded files.
- Decided on your preferred installation method based on available resources and project requirements.
- Pre-Installation Checklist:
- Verify that your system meets the hardware and software requirements outlined in Chapter 2.
- Ensure all required dependencies, including Python, pip, Git, and CUDA (if applicable), are installed.
- Downloaded ESM3 Repository:
- Ensure the ESM3 repository has been downloaded or cloned, as detailed in Chapter 3.
- Environment Setup:
- Activate the Python virtual environment created in Chapter 2:
source esm3_env/bin/activate
- Activate the Python virtual environment created in Chapter 2:
- Move into the directory where the ESM3 repository was downloaded or cloned:
cd ~/esm3_workspace/esm - Use
pipto install the required Python libraries:pip install -r requirements.txt - If you plan to use GPU acceleration, install the CUDA Toolkit and cuDNN, as detailed in Chapter 2. Confirm GPU availability with:
nvidia-smi - Install ESM3 by running the setup script:
python setup.py install - Test the installation by running a basic ESM3 command:
python examples/run_pretrained_model.py --help - Move to the directory containing the ESM3 repository:
cd ~/esm3_workspace/esm - Use Homebrew to install system-level dependencies:
brew install cmake - Install Python dependencies with
pip:pip install -r requirements.txt - Run the installation script:
python setup.py install - Test ESM3 functionality with:
python examples/run_pretrained_model.py --help - Install WSL2 and a Linux distribution (e.g., Ubuntu):powershellCopy code
wsl --install - Launch the Linux terminal and update the system:
sudo apt update && sudo apt upgrade -y - Follow the Linux installation instructions to set up Python, pip, Git, and other required tools:
sudo apt install python3 python3-pip git build-essential cmake - Follow NVIDIA’s official instructions to install WSL-compatible CUDA drivers.
- Change to the directory where the ESM3 repository was cloned:
cd ~/esm3_workspace/esm - Install ESM3 using the setup script:
python setup.py install - Test ESM3 functionality:
python examples/run_pretrained_model.py --help - Run the Help Command:
- Execute a basic command to display help options:
python examples/run_pretrained_model.py --help
- Execute a basic command to display help options:
- Test with Sample Data:
- Run a test using sample input data:
python examples/run_pretrained_model.py esm2_t6_8M_UR50D data/sample.fasta - Verify that the output includes predicted protein structures and confidence scores.
- Run a test using sample input data:
- Check GPU Utilization (if applicable):
- Ensure GPU acceleration is functioning:
nvidia-smi
- Ensure GPU acceleration is functioning:
- Resolve Issues:
- If errors occur, consult the troubleshooting guide in Chapter 7.
- Update Environment Variables:
- Add the ESM3 directory to your PATH for easier command execution:
export PATH= PATH:~/esm3_workspace/esm
PATH:~/esm3_workspace/esm - Add this line to your shell configuration file (e.g.,
.bashrcor.zshrc) to make it persistent:echo 'export PATH= PATH
PATH - Test access to ESM3 commands:
run_pretrained_model.py --help
5.2 Enabling GPU Acceleration
GPU acceleration dramatically improves ESM3’s performance, especially for large datasets. Proper configuration ensures that the tool fully utilizes your system’s GPU capabilities.
Step 1: Verify GPU and CUDA Installation
- Check if your system has a compatible GPU:
nvidia-smi - Ensure that the CUDA Toolkit and cuDNN are installed and compatible with your GPU:
nvcc --version
Step 2: Install Required Libraries
- Install PyTorch with GPU support, as it is a key dependency for ESM3:
pip install torch torchvision torchaudio --index-url https://download.pytorch.org/whl/cu118
Step 3: Configure ESM3 for GPU Use
- Modify the configuration file to specify GPU usage:
- Locate the configuration file (if applicable) or create a runtime argument for GPU:
--device cuda
- Locate the configuration file (if applicable) or create a runtime argument for GPU:
- Test GPU functionality by running a sample command:
python examples/run_pretrained_model.py esm2_t6_8M_UR50D --device cuda data/sample.fasta
5.3 Customizing Default Configurations
Customizing ESM3 configurations allows you to optimize workflows and adapt the tool to your specific research needs.
Step 1: Adjust Default Parameters
- Locate and edit configuration files (if provided) or set parameters directly in the command line:
- Batch Size: Adjust for memory constraints:
--batch_size 32 - Output Format: Specify desired output (e.g., JSON, CSV):
--output_format json
- Batch Size: Adjust for memory constraints:
Step 2: Set Input and Output Paths
- Define default directories for input sequences and output files:
export ESM3_INPUT_DIR=~/esm3_workspace/inputs export ESM3_OUTPUT_DIR=~/esm3_workspace/outputs - Update your shell configuration file for persistence:
echo 'export ESM3_INPUT_DIR=~/esm3_workspace/inputs' >> ~/.bashrc echo 'export ESM3_OUTPUT_DIR=~/esm3_workspace/outputs' >> ~/.bashrc source ~/.bashrc
Step 3: Automate Workflows
- Create reusable scripts for frequently used commands:
echo 'python examples/run_pretrained_model.py esm2_t6_8M_UR50D --device cuda file done
file done
2. Multi-GPU Configuration
- If using multiple GPUs, configure ESM3 to distribute workloads:
torch.distributed.init_process_group(backend="nccl")
3. Cloud and Cluster Configurations
- For users deploying ESM3 on cloud platforms or HPC clusters:
- Set up job scheduling for batch predictions (e.g., SLURM):
sbatch run_esm3_job.sh - Use cloud-native solutions like Google Colab or AWS to bypass local resource constraints.
- Set up job scheduling for batch predictions (e.g., SLURM):
5.5 Verifying Configuration
After making changes, verify that the configurations are applied correctly:
- Run a Test Command:
- Use sample input data to confirm the configuration works:
python examples/run_pretrained_model.py esm2_t6_8M_UR50D --device cuda data/sample.fasta
- Use sample input data to confirm the configuration works:
- Check Resource Utilization:
- Monitor GPU or CPU usage during execution:
nvidia-smi htop
- Monitor GPU or CPU usage during execution:
- Validate Outputs:
- Ensure that output files are generated in the specified format and directory:
ls ~/esm3_workspace/outputs
- Ensure that output files are generated in the specified format and directory:
5.6 Preparing for Workflow Integration
Proper configuration ensures ESM3 is ready for integration into larger workflows:
- Interfacing with Other Tools:
- Connect ESM3 outputs to visualization tools like PyMOL or Chimera for structure analysis.
- Linking with Automation Pipelines:
- Use tools like Snakemake or Nextflow to create automated workflows that include ESM3.
Configuring ESM3 is a critical step to ensure optimal performance and seamless integration into your research workflows. From enabling GPU acceleration to customizing input and output settings, the steps outlined in this chapter provide the foundation for a flexible and efficient setup. With configurations in place, the next chapter will guide you through running ESM3 for the first time and interpreting its outputs effectively.
6. Running ESM3 for the First Time
After successfully installing and configuring ESM3 (Evolutionary Scale Modeling 3), the next step is to run the software and interpret its outputs. This chapter provides a detailed, step-by-step guide to executing ESM3 for the first time, using sample data to test functionality and ensure that the tool is working as expected. Additionally, it covers best practices for input formatting, running predictions, and analyzing outputs.
6.1 Preparing Input Data
ESM3 requires input data in specific formats, most commonly FASTA files for protein sequences. Properly formatted input ensures accurate predictions and prevents runtime errors.
Step 1: Understand FASTA Format
- A FASTA file consists of protein sequences in the following format:shellCopy code
>sequence_id MTEYKLVVVGAGGVGKSALTIQLIQNHFVDEYDPTIEDSYRKQ - Each sequence must have a unique identifier preceded by a
>symbol, followed by the amino acid sequence on the next line.
Step 2: Obtain Sample Data
- Download example FASTA files from the official ESM3 GitHub repository or prepare your own sequences:
wget https://github.com/facebookresearch/esm/raw/main/examples/sample.fasta
Step 3: Validate Input Data
- Check the integrity and formatting of the input file:
head -n 10 sample.fasta - Ensure there are no special characters or spaces in the sequence lines.
Step 4: Save Input in a Designated Directory
- Place your input file in the directory specified during configuration (e.g.,
~/esm3_workspace/inputs).
6.2 Running a Basic Prediction
The simplest way to test ESM3 is by running a basic prediction using pre-trained models.
Step 1: Navigate to the ESM3 Directory
- Move into the ESM3 installation directory:
cd ~/esm3_workspace/esm
Step 2: Execute the Prediction Command
- Use the following command to run a prediction:
python examples/run_pretrained_model.py esm2_t6_8M_UR50D data/sample.fasta - Replace
esm2_t6_8M_UR50Dwith the desired pre-trained model anddata/sample.fastawith the path to your input file.
Step 3: Monitor Execution
- During execution, ESM3 will:
- Load the pre-trained model.
- Process the input sequences.
- Generate outputs, including predictions and confidence scores.
Step 4: Review Outputs
- After the command completes, check the output directory (e.g.,
~/esm3_workspace/outputs) for results:ls ~/esm3_workspace/outputs - Common output files include:
- Predicted structure files (e.g., PDB or PyTorch tensors).
- Confidence scores (e.g., CSV files).
6.3 Understanding ESM3 Output
The outputs generated by ESM3 provide valuable insights into protein structure and function.
1. Structural Predictions
- Output: Predicted 3D coordinates of the protein structure in formats such as PDB or PyTorch tensors.
- Applications:
- Visualize the structure using molecular visualization tools like PyMOL or Chimera:
pymol ~/esm3_workspace/outputs/sample.pdb
- Visualize the structure using molecular visualization tools like PyMOL or Chimera:
2. Confidence Scores
- Output: A CSV file containing confidence scores for each residue in the predicted structure.
- Applications:
- Use confidence scores to identify regions with high structural reliability.
- Example CSV content:Copy code
residue_id,confidence_score 1,0.85 2,0.78 3,0.92
3. Sequence Annotations
- Output: Functional annotations or predicted domains (if applicable, based on the model used).
- Applications:
- Analyze functional sites such as ligand-binding regions or active sites.
6.4 Troubleshooting First-Time Runs
If issues arise during the first run, consider these troubleshooting steps:
1. Common Errors
- Error: Missing Dependencies
- Ensure all required Python libraries are installed:
pip install -r requirements.txt
- Ensure all required Python libraries are installed:
- Error: CUDA Not Available
- Verify GPU compatibility and installation:
nvidia-smi
- Verify GPU compatibility and installation:
- Error: Invalid Input File
- Check input file formatting for errors:
cat data/sample.fasta
- Check input file formatting for errors:
2. Debugging Tips
- Run the command with a debugging flag (if available) to identify issues:
python examples/run_pretrained_model.py esm2_t6_8M_UR50D data/sample.fasta --debug - Consult the ESM3 GitHub repository for known issues and solutions.
6.5 Best Practices for First-Time Runs
To ensure a successful first run, follow these best practices:
- Start Small:
- Use small input files with a limited number of sequences to test the setup before processing larger datasets.
- Check Outputs Immediately:
- Validate that output files are complete and correctly formatted.
- Document Results:
- Maintain a log of commands run and their outputs for future reference.
- Monitor Resource Usage:
- Use tools like
nvidia-smi(GPU) orhtop(CPU) to ensure efficient resource utilization.
- Use tools like
- Verify Model Selection:
- Choose the appropriate pre-trained model based on your research goals.
6.6 Preparing for Advanced Workflows
Once the basic prediction is successful, you can prepare for more advanced workflows:
- Batch Processing:
- Automate predictions for multiple input files using shell scripts.
- Example:
for file in ~/esm3_workspace/inputs/*.fasta; do python examples/run_pretrained_model.py esm2_t6_8M_UR50D file done
file done - Step 3: Combine outputs:
- Merge output files from all batches into a single file:
cat outputs/batch_* > combined_output.csv
- Merge output files from all batches into a single file:
2. Parallel Processing
- Utilize multiple CPU cores or GPUs to process data simultaneously:
- Example: Using GNU Parallel for multi-threaded execution:
parallel python examples/run_pretrained_model.py esm2_t6_8M_UR50D --device cuda ::: batch_*
- Example: Using GNU Parallel for multi-threaded execution:
7.2 Multi-GPU Configuration
For users with access to multiple GPUs, configuring ESM3 to distribute workloads across GPUs can significantly enhance performance.
1. Enable Multi-GPU Mode
- Modify the runtime arguments to specify multiple devices:
python examples/run_pretrained_model.py esm2_t6_8M_UR50D --device cuda:0,cuda:1 data/sample.fasta
2. Adjust Batch Size
- Divide the input sequences across GPUs by adjusting the batch size:
--batch_size 64
3. Test Multi-GPU Configuration
- Monitor GPU usage to verify that both GPUs are utilized:
nvidia-smi
7.3 Automating Workflows
Automating ESM3 workflows reduces manual effort and ensures consistency across multiple runs.
1. Create Shell Scripts
- Example shell script for running ESM3 on a list of files:
#!/bin/bash for file in ~/esm3_workspace/inputs/*.fasta; do python examples/run_pretrained_model.py esm2_t6_8M_UR50D --device cuda![Rendered by QuickLaTeX.com file done</code></li> <!-- /wp:list-item --> <!-- wp:list-item --> <li>Save the script as <code>run_esm3.sh</code> and make it executable: <code>chmod +x run_esm3.sh ./run_esm3.sh</code></li> <!-- /wp:list-item --></ul> <!-- /wp:list --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>2. Automate Output Analysis</strong></h4> <!-- /wp:heading --> <!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Post-process outputs with Python or shell scripts:<!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Example: Extract high-confidence regions:pythonCopy code<code>import pandas as pd data = pd.read_csv("combined_output.csv") high_conf = data[data["confidence_score"] > 0.9] high_conf.to_csv("high_confidence_regions.csv", index=False)</code></li> <!-- /wp:list-item --></ul> <!-- /wp:list --></li> <!-- /wp:list-item --></ul> <!-- /wp:list --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:heading {"level":3} --> <h3 class="wp-block-heading"><strong>7.4 Integrating with Cloud Platforms</strong></h3> <!-- /wp:heading --> <!-- wp:paragraph --> Cloud computing provides scalability and flexibility, enabling ESM3 to handle computationally intensive tasks without requiring local resources. <!-- /wp:paragraph --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>1. Using Google Colab</strong></h4> <!-- /wp:heading --> <!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Google Colab provides free GPU resources for running ESM3:<!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Open a Colab notebook and install ESM3:pythonCopy code<code>!git clone https://github.com/facebookresearch/esm.git !pip install -r esm/requirements.txt</code></li> <!-- /wp:list-item --> <!-- wp:list-item --> <li>Run predictions:pythonCopy code<code>!python esm/examples/run_pretrained_model.py esm2_t6_8M_UR50D esm/examples/sample.fasta</code></li> <!-- /wp:list-item --></ul> <!-- /wp:list --></li> <!-- /wp:list-item --></ul> <!-- /wp:list --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>2. Deploying on AWS</strong></h4> <!-- /wp:heading --> <!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Use an EC2 instance with GPU capabilities:<!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Select an instance type (e.g., <strong>g4dn.xlarge</strong>) and install required libraries: <code>sudo apt update && sudo apt install python3 python3-pip git -y pip install torch torchvision torchaudio --index-url https://download.pytorch.org/whl/cu118</code></li> <!-- /wp:list-item --> <!-- wp:list-item --> <li>Clone the ESM3 repository and run predictions.</li> <!-- /wp:list-item --></ul> <!-- /wp:list --></li> <!-- /wp:list-item --></ul> <!-- /wp:list --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>3. Leveraging HPC Clusters</strong></h4> <!-- /wp:heading --> <!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>For large-scale analyses, deploy ESM3 on high-performance computing (HPC) clusters:<!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Create a SLURM batch script: <code>#!/bin/bash #SBATCH --job-name=esm3_job #SBATCH --gres=gpu:1 #SBATCH --cpus-per-task=4 #SBATCH --time=01:00:00 module load python source esm3_env/bin/activate python examples/run_pretrained_model.py esm2_t6_8M_UR50D data/sample.fasta</code></li> <!-- /wp:list-item --> <!-- wp:list-item --> <li>Submit the job: <code>sbatch run_esm3_job.sh</code></li> <!-- /wp:list-item --></ul> <!-- /wp:list --></li> <!-- /wp:list-item --></ul> <!-- /wp:list --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:heading {"level":3} --> <h3 class="wp-block-heading"><strong>7.5 Workflow Integration with Bioinformatics Tools</strong></h3> <!-- /wp:heading --> <!-- wp:paragraph --> Integrating ESM3 with other bioinformatics tools enhances its functionality and streamlines complex workflows. <!-- /wp:paragraph --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>1. Visualization Tools</strong></h4> <!-- /wp:heading --> <!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Use outputs from ESM3 in molecular visualization tools:<!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>PyMOL: <code>pymol outputs/sample.pdb</code></li> <!-- /wp:list-item --> <!-- wp:list-item --> <li>Chimera: <code>chimera --script outputs/sample.pdb</code></li> <!-- /wp:list-item --></ul> <!-- /wp:list --></li> <!-- /wp:list-item --></ul> <!-- /wp:list --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>2. Downstream Analysis</strong></h4> <!-- /wp:heading --> <!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Combine ESM3 predictions with molecular docking or dynamics simulations:<!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Docking: Use predicted structures as inputs for docking simulations in tools like AutoDock or Rosetta.</li> <!-- /wp:list-item --> <!-- wp:list-item --> <li>Molecular Dynamics: Refine ESM3-generated models using MD simulations in GROMACS or Amber.</li> <!-- /wp:list-item --></ul> <!-- /wp:list --></li> <!-- /wp:list-item --></ul> <!-- /wp:list --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>3. Automating Pipelines</strong></h4> <!-- /wp:heading --> <!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Use workflow management tools like Snakemake or Nextflow:<!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Example Snakemake rule:yamlCopy code<code>rule run_esm3: input: "inputs/{file}.fasta" output: "outputs/{file}.pdb" shell: "python examples/run_pretrained_model.py esm2_t6_8M_UR50D --device cuda {input} > {output}"</code></li> <!-- /wp:list-item --></ul> <!-- /wp:list --></li> <!-- /wp:list-item --></ul> <!-- /wp:list --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:heading {"level":3} --> <h3 class="wp-block-heading"><strong>7.6 Testing and Validating Configurations</strong></h3> <!-- /wp:heading --> <!-- wp:paragraph --> After implementing advanced configurations, test them thoroughly to ensure functionality and performance: <!-- /wp:paragraph --> <!-- wp:list {"ordered":true} --> <ol class="wp-block-list"><!-- wp:list-item --> <li><strong>Run Test Commands:</strong><!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Verify that batch processing, multi-GPU setups, and automation scripts work as intended.</li> <!-- /wp:list-item --></ul> <!-- /wp:list --></li> <!-- /wp:list-item --> <!-- wp:list-item --> <li><strong>Monitor Resource Utilization:</strong><!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Check CPU and GPU usage during execution to optimize resource allocation: <code>htop nvidia-smi</code></li> <!-- /wp:list-item --></ul> <!-- /wp:list --></li> <!-- /wp:list-item --> <!-- wp:list-item --> <li><strong>Validate Outputs:</strong><!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Confirm that output files are complete and correctly formatted for downstream analysis.</li> <!-- /wp:list-item --></ul> <!-- /wp:list --></li> <!-- /wp:list-item --></ol> <!-- /wp:list --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:paragraph --> Advanced configurations enable users to scale ESM3 for high-throughput workflows, leverage cloud computing, and integrate it with complementary bioinformatics tools. By following the detailed instructions provided in this chapter, researchers can enhance ESM3's efficiency and functionality, tailoring it to the demands of complex projects. The next chapter will address common troubleshooting scenarios, providing solutions to potential issues encountered during installation, configuration, and execution. <!-- /wp:paragraph --> <!-- wp:heading {"level":3} --> <h3 class="wp-block-heading"><strong>8. Troubleshooting Common Issues</strong></h3> <!-- /wp:heading --> <!-- wp:paragraph --> While <strong>ESM3 (Evolutionary Scale Modeling 3)</strong> is designed to be robust and accessible, users may encounter challenges during installation, configuration, or execution. Addressing these issues promptly ensures uninterrupted research workflows. This chapter provides a comprehensive troubleshooting guide, covering common errors, diagnostic techniques, and solutions for potential problems. <!-- /wp:paragraph --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:heading {"level":3} --> <h3 class="wp-block-heading"><strong>8.1 Installation Issues</strong></h3> <!-- /wp:heading --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>Problem: Missing or Incompatible Dependencies</strong></h4> <!-- /wp:heading --> <!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li><strong>Symptom:</strong> Errors such as <code>ModuleNotFoundError</code> or <code>Incompatible library version</code> during installation or execution.</li> <!-- /wp:list-item --> <!-- wp:list-item --> <li><strong>Diagnosis:</strong> Missing or outdated Python libraries, or incompatible versions of tools like PyTorch, CMake, or CUDA.</li> <!-- /wp:list-item --> <!-- wp:list-item --> <li><strong>Solution:</strong><!-- wp:list {"ordered":true} --> <ol class="wp-block-list"><!-- wp:list-item --> <li>Verify the Python environment: <code>python3 --version</code></li> <!-- /wp:list-item --> <!-- wp:list-item --> <li>Reinstall dependencies: <code>pip install -r requirements.txt --upgrade</code></li> <!-- /wp:list-item --> <!-- wp:list-item --> <li>Ensure CUDA compatibility with PyTorch: <code>python -c "import torch; print(torch.cuda.is_available())"</code></li> <!-- /wp:list-item --></ol> <!-- /wp:list --></li> <!-- /wp:list-item --></ul> <!-- /wp:list --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>Problem: Incorrect System Configuration</strong></h4> <!-- /wp:heading --> <!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li><strong>Symptom:</strong> Installation fails or ESM3 commands are not recognized.</li> <!-- /wp:list-item --> <!-- wp:list-item --> <li><strong>Diagnosis:</strong> PATH variable not updated or incomplete setup.</li> <!-- /wp:list-item --> <!-- wp:list-item --> <li><strong>Solution:</strong><!-- wp:list {"ordered":true} --> <ol class="wp-block-list"><!-- wp:list-item --> <li>Confirm PATH configuration: <code>echo](https://esm3academy.com/wp-content/ql-cache/quicklatex.com-3a0d08cd7662263cdd87ef02b18bf496_l3.png) PATH
PATH - Add ESM3 directory to the PATH:
export PATH= ESM3_OUTPUT_DIR
ESM3_OUTPUT_DIR - Update paths:
export ESM3_OUTPUT_DIR=~/esm3_workspace/outputs source ~/.bashrc
- Add the ESM3 directory to your PATH for easier command execution:
- Symptom: Error messages such as
Invalid FASTA formatorUnrecognized input file. - Diagnosis: Input file contains formatting errors or unsupported sequences.
- Solution:
- Validate input file formatting:
head -n 10 data/sample.fasta - Ensure proper FASTA format:shellCopy code
>sequence_id MTEYKLVVVGAGGVGKSALTIQLIQNHFVDEYDPTIEDSYRKQ - Use a sequence validator tool to check for errors.
- Validate input file formatting:
- Symptom: Execution fails with
Out of memoryerror. - Diagnosis: Input file size or batch size exceeds available RAM or GPU memory.
- Solution:
- Reduce batch size:
--batch_size 32 - Split large input files into smaller batches:
split -l 1000 large_input.fasta batch_ - Use a smaller pre-trained model if possible.
- Reduce batch size:
- Symptom: ESM3 processes data much slower than expected.
- Diagnosis: Suboptimal resource utilization or CPU-only execution.
- Solution:
- Ensure GPU is being used:
python -c "import torch; print(torch.cuda.is_available())" - Monitor resource usage:
nvidia-smi htop - Optimize resource allocation by using advanced configurations such as parallel processing (Chapter 7).
- Ensure GPU is being used:
- Symptom: Disk space fills up quickly during execution.
- Diagnosis: Temporary files or large outputs are not managed properly.
- Solution:
- Clean up temporary files after execution:
rm -rf ~/esm3_workspace/tmp/* - Use external storage for large output files.
- Clean up temporary files after execution:
- Symptom: Output directory is empty, or files cannot be opened.
- Diagnosis: Execution failed partway through or output directory permissions are incorrect.
- Solution:
- Check log files for errors:
cat logs/run_esm3.log - Verify output directory permissions:
chmod 755 ~/esm3_workspace/outputs - Re-run the prediction on a smaller dataset to isolate issues.
- Check log files for errors:
- Symptom: Predicted structures or confidence scores seem incorrect.
- Diagnosis: Input sequences may not align with the model’s training set, or the wrong model was used.
- Solution:
- Verify the pre-trained model matches the input data:
esm2_t6_8M_UR50D vs esm2_t33_650M_UR50D - Test with a known dataset to validate model behavior.
- Verify the pre-trained model matches the input data:
- Refer to the ESM3 GitHub repository for detailed installation and usage instructions:
https://github.com/facebookresearch/esm. - Join discussion forums and GitHub Issues for troubleshooting advice from other users and developers.
- Use logging flags for detailed output during execution:
python examples/run_pretrained_model.py esm2_t6_8M_UR50D data/sample.fasta --debug - Regularly check the official ESM3 GitHub repository for new releases: https://github.com/facebookresearch/esm.
- If you cloned the repository during installation, update it periodically:
cd ~/esm3_workspace/esm git pull origin main - Some updates may introduce new dependencies. Reinstall requirements to ensure compatibility:
pip install -r requirements.txt --upgrade - Run a small test to verify that ESM3 works as expected after updates:
python examples/run_pretrained_model.py esm2_t6_8M_UR50D data/sample.fasta - Use scripts to streamline repetitive tasks like batch processing:
for file in ~/esm3_workspace/inputs/*.fasta; do python examples/run_pretrained_model.py esm2_t6_8M_UR50D --device cuda![Rendered by QuickLaTeX.com file done</code></li> <!-- /wp:list-item --></ul> <!-- /wp:list --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>2. Use Scalable Configurations</strong></h4> <!-- /wp:heading --> <!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Adjust batch sizes based on available memory: <code>--batch_size 64</code></li> <!-- /wp:list-item --> <!-- wp:list-item --> <li>Deploy workflows on scalable cloud platforms or HPC clusters for larger datasets.</li> <!-- /wp:list-item --></ul> <!-- /wp:list --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>3. Optimize Resource Allocation</strong></h4> <!-- /wp:heading --> <!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Monitor resource usage to ensure efficient allocation:<!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Use <code>nvidia-smi</code> for GPU monitoring.</li> <!-- /wp:list-item --> <!-- wp:list-item --> <li>Use <code>htop</code> or <code>top</code> for CPU and memory tracking.</li> <!-- /wp:list-item --></ul> <!-- /wp:list --></li> <!-- /wp:list-item --></ul> <!-- /wp:list --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:heading {"level":3} --> <h3 class="wp-block-heading"><strong>9.3 Maintaining a Stable Environment</strong></h3> <!-- /wp:heading --> <!-- wp:paragraph --> A stable computational environment reduces the risk of runtime errors and ensures reproducibility in research. <!-- /wp:paragraph --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>1. Use Virtual Environments</strong></h4> <!-- /wp:heading --> <!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Always activate the Python virtual environment before running ESM3: <code>source esm3_env/bin/activate</code></li> <!-- /wp:list-item --></ul> <!-- /wp:list --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>2. Archive Working Configurations</strong></h4> <!-- /wp:heading --> <!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Save copies of configuration files and scripts that work well for your workflows: <code>cp esm3_config.py esm3_config_backup.py</code></li> <!-- /wp:list-item --></ul> <!-- /wp:list --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>3. Document Changes</strong></h4> <!-- /wp:heading --> <!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Maintain a log of updates, workflow adjustments, and command-line options for reference in future projects.</li> <!-- /wp:list-item --></ul> <!-- /wp:list --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:heading {"level":3} --> <h3 class="wp-block-heading"><strong>9.4 Managing Outputs</strong></h3> <!-- /wp:heading --> <!-- wp:paragraph --> Efficiently organizing and storing outputs ensures easy access and minimizes the risk of data loss. <!-- /wp:paragraph --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>1. Organize Outputs</strong></h4> <!-- /wp:heading --> <!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Use a consistent directory structure to store results: <code>~/esm3_workspace/outputs/ ├── batch1/ ├── batch2/ └── combined_results/</code></li> <!-- /wp:list-item --></ul> <!-- /wp:list --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>2. Backup Results</strong></h4> <!-- /wp:heading --> <!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Regularly back up output files to external storage or cloud services: <code>rsync -av ~/esm3_workspace/outputs/ /path/to/backup/</code></li> <!-- /wp:list-item --></ul> <!-- /wp:list --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>3. Use Output Analysis Tools</strong></h4> <!-- /wp:heading --> <!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Automate post-processing of outputs for downstream analysis:pythonCopy code<code>import pandas as pd data = pd.read_csv("combined_output.csv") high_conf = data[data["confidence_score"] > 0.9] high_conf.to_csv("high_confidence_regions.csv", index=False)</code></li> <!-- /wp:list-item --></ul> <!-- /wp:list --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:heading {"level":3} --> <h3 class="wp-block-heading"><strong>9.5 Adapting to New Use Cases</strong></h3> <!-- /wp:heading --> <!-- wp:paragraph --> As research needs change, adapting ESM3 to new use cases ensures continued relevance and utility. <!-- /wp:paragraph --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>1. Experiment with New Pre-Trained Models</strong></h4> <!-- /wp:heading --> <!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>New models are regularly added to the ESM3 repository. Test these models for specific applications: <code>python examples/run_pretrained_model.py esm2_t33_650M_UR50D data/sample.fasta</code></li> <!-- /wp:list-item --></ul> <!-- /wp:list --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>2. Integrate with Emerging Tools</strong></h4> <!-- /wp:heading --> <!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Link ESM3 outputs to newer bioinformatics or computational tools:<!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Example: Feed predicted structures into molecular docking simulations or functional annotation tools.</li> <!-- /wp:list-item --></ul> <!-- /wp:list --></li> <!-- /wp:list-item --></ul> <!-- /wp:list --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>3. Participate in the Community</strong></h4> <!-- /wp:heading --> <!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Join discussions, report issues, and contribute to the ESM3 community: <a href="https://github.com/facebookresearch/esm/issues">https://github.com/facebookresearch/esm/issues</a>.</li> <!-- /wp:list-item --></ul> <!-- /wp:list --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:heading {"level":3} --> <h3 class="wp-block-heading"><strong>9.6 Ensuring Reproducibility</strong></h3> <!-- /wp:heading --> <!-- wp:paragraph --> Reproducibility is essential for collaborative research and validation of results. <!-- /wp:paragraph --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>1. Version Control for Code and Data</strong></h4> <!-- /wp:heading --> <!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Use Git to track changes in scripts and configuration files: <code>git init git add . git commit -m "Initial setup for ESM3 workflow"</code></li> <!-- /wp:list-item --></ul> <!-- /wp:list --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>2. Record Metadata</strong></h4> <!-- /wp:heading --> <!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Log metadata for each analysis, including:<!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Input file names.</li> <!-- /wp:list-item --> <!-- wp:list-item --> <li>Model versions used.</li> <!-- /wp:list-item --> <!-- wp:list-item --> <li>Command-line options.</li> <!-- /wp:list-item --></ul> <!-- /wp:list --></li> <!-- /wp:list-item --></ul> <!-- /wp:list --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>3. Share Reproducible Pipelines</strong></h4> <!-- /wp:heading --> <!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Package workflows using tools like Snakemake or Docker for easy sharing:<!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Example Dockerfile for ESM3:DockerfileCopy code<code>FROM python:3.8 RUN git clone https://github.com/facebookresearch/esm.git WORKDIR /esm RUN pip install -r requirements.txt CMD ["bash"]</code></li> <!-- /wp:list-item --></ul> <!-- /wp:list --></li> <!-- /wp:list-item --></ul> <!-- /wp:list --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:heading {"level":3} --> <h3 class="wp-block-heading"><strong>9.7 Long-Term Maintenance Tips</strong></h3> <!-- /wp:heading --> <!-- wp:paragraph --> To ensure ESM3 remains functional and efficient over time, adopt these practices: <!-- /wp:paragraph --> <!-- wp:list {"ordered":true} --> <ol class="wp-block-list"><!-- wp:list-item --> <li><strong>Monitor System Health:</strong><!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Regularly update system libraries and drivers: <code>sudo apt update && sudo apt upgrade -y</code></li> <!-- /wp:list-item --></ul> <!-- /wp:list --></li> <!-- /wp:list-item --> <!-- wp:list-item --> <li><strong>Plan for Hardware Upgrades:</strong><!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Consider upgrading GPUs or adding more RAM for handling increasingly complex analyses.</li> <!-- /wp:list-item --></ul> <!-- /wp:list --></li> <!-- /wp:list-item --> <!-- wp:list-item --> <li><strong>Follow ESM3 Developments:</strong><!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Stay informed about new features, models, and use cases by monitoring the repository and associated publications.</li> <!-- /wp:list-item --></ul> <!-- /wp:list --></li> <!-- /wp:list-item --></ol> <!-- /wp:list --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:paragraph --> Maintaining ESM3 for long-term use requires a combination of regular updates, workflow optimization, and proactive management of outputs and configurations. By following the best practices outlined in this chapter, users can ensure consistent performance, adaptability to new research challenges, and reproducibility of results. With ESM3 configured and maintained, you are now equipped to fully leverage its capabilities for advanced protein modeling and related applications. <!-- /wp:paragraph --> <!-- wp:heading {"level":3} --> <h3 class="wp-block-heading"><strong>10. Integrating ESM3 into Complex Workflows</strong></h3> <!-- /wp:heading --> <!-- wp:paragraph --> For advanced users, integrating <strong>ESM3 (Evolutionary Scale Modeling 3)</strong> into complex workflows allows for seamless interaction with complementary tools and technologies. This chapter explores strategies to incorporate ESM3 into pipelines for large-scale bioinformatics projects, molecular simulations, and automated systems. By creating interoperable workflows, researchers can maximize ESM3's utility across various scientific domains. <!-- /wp:paragraph --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:heading {"level":3} --> <h3 class="wp-block-heading"><strong>10.1 Designing Interoperable Pipelines</strong></h3> <!-- /wp:heading --> <!-- wp:paragraph --> Integration begins with designing pipelines that incorporate ESM3 as a modular component for data processing, prediction, and downstream analysis. <!-- /wp:paragraph --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>1. Workflow Management Tools</strong></h4> <!-- /wp:heading --> <!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Leverage tools like <strong>Snakemake</strong> or <strong>Nextflow</strong> to structure and automate multi-step workflows.</li> <!-- /wp:list-item --> <!-- wp:list-item --> <li>Example Snakemake Rule for ESM3:yamlCopy code<code>rule run_esm3: input: "inputs/{sequence_file}.fasta" output: "outputs/{sequence_file}.pdb" shell: "python examples/run_pretrained_model.py esm2_t6_8M_UR50D --device cuda {input} > {output}"</code></li> <!-- /wp:list-item --></ul> <!-- /wp:list --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>2. Modular Workflow Design</strong></h4> <!-- /wp:heading --> <!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Divide workflows into stages:<!-- wp:list {"ordered":true} --> <ol class="wp-block-list"><!-- wp:list-item --> <li>Data preprocessing (e.g., sequence cleaning).</li> <!-- /wp:list-item --> <!-- wp:list-item --> <li>Prediction using ESM3.</li> <!-- /wp:list-item --> <!-- wp:list-item --> <li>Post-processing and downstream analysis.</li> <!-- /wp:list-item --></ol> <!-- /wp:list --></li> <!-- /wp:list-item --></ul> <!-- /wp:list --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>3. Integrate with Other Tools</strong></h4> <!-- /wp:heading --> <!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Use intermediate outputs from ESM3 as inputs for other tools:<!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li><strong>Functional Annotation:</strong> Feed predicted structures into tools like InterProScan.</li> <!-- /wp:list-item --> <!-- wp:list-item --> <li><strong>Molecular Docking:</strong> Use docking tools such as AutoDock or Rosetta.</li> <!-- /wp:list-item --></ul> <!-- /wp:list --></li> <!-- /wp:list-item --></ul> <!-- /wp:list --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:heading {"level":3} --> <h3 class="wp-block-heading"><strong>10.2 Preprocessing Input Data</strong></h3> <!-- /wp:heading --> <!-- wp:paragraph --> Properly formatted input ensures accuracy and compatibility across tools. <!-- /wp:paragraph --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>1. Sequence Validation</strong></h4> <!-- /wp:heading --> <!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Use FASTA validators to ensure sequences conform to accepted standards: <code>fasta-validator data/sample.fasta</code></li> <!-- /wp:list-item --></ul> <!-- /wp:list --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>2. Filtering Redundant Sequences</strong></h4> <!-- /wp:heading --> <!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Remove duplicate or highly similar sequences to optimize processing: <code>cd-hit -i data/sample.fasta -o data/unique_sequences.fasta -c 0.9</code></li> <!-- /wp:list-item --></ul> <!-- /wp:list --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>3. Custom Annotations</strong></h4> <!-- /wp:heading --> <!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Add functional annotations to sequences for context:shellCopy code<code>>sequence_id|function=kinase MTEYKLVVVGAGGVGKSALTIQLIQNHFVDEYDPTIEDSYRKQ</code></li> <!-- /wp:list-item --></ul> <!-- /wp:list --> <!-- wp:separator --> <hr class="wp-block-separator has-alpha-channel-opacity"/> <!-- /wp:separator --> <!-- wp:heading {"level":3} --> <h3 class="wp-block-heading"><strong>10.3 Automating Batch Processing</strong></h3> <!-- /wp:heading --> <!-- wp:paragraph --> Large-scale projects often require processing thousands of sequences. Automating batch workflows minimizes manual effort. <!-- /wp:paragraph --> <!-- wp:heading {"level":4} --> <h4 class="wp-block-heading"><strong>1. Shell Script for Batch Predictions</strong></h4> <!-- /wp:heading --> <!-- wp:list --> <ul class="wp-block-list"><!-- wp:list-item --> <li>Example script to automate batch processing: <code>for file in ~/esm3_workspace/inputs/*.fasta; do python examples/run_pretrained_model.py esm2_t6_8M_UR50D --device cuda](https://esm3academy.com/wp-content/ql-cache/quicklatex.com-0a0ec9589dc603da3815592b23c39338_l3.png) file > outputs/
file > outputs/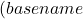 file .fasta).pdb done
file .fasta).pdb done - Use GNU Parallel for concurrent processing:
parallel python examples/run_pretrained_model.py esm2_t6_8M_UR50D --device cuda ::: ~/esm3_workspace/inputs/*.fasta - Convert predicted structures to compatible formats:
obabel sample.pdb -O sample.gro - Use tools like GROMACS or AMBER for simulation:
- Example GROMACS Workflow:
gmx pdb2gmx -f sample.pdb -o processed.gro -water spce gmx editconf -f processed.gro -o box.gro -c -d 1.0 -bt cubic gmx grompp -f em.mdp -c box.gro -p topol.top -o em.tpr gmx mdrun -v -deffnm em
- Example GROMACS Workflow:
- Evaluate the stability of refined structures using root-mean-square deviation (RMSD):
gmx rms -s em.tpr -f traj.xtc -o rmsd.xvg - Integrate ESM3 predictions with annotation tools like InterProScan:
interproscan.sh -i outputs/sample.pdb -o annotations.tsv - Prepare ESM3-predicted structures for docking:
- Add hydrogens and remove water molecules:
obabel sample.pdb -h -O sample_hydrogenated.pdb
- Add hydrogens and remove water molecules:
- Run docking using AutoDock or similar tools:
vina --receptor sample_hydrogenated.pdb --ligand ligand.pdb --out docking_results.pdb - Use ESM3 outputs to predict and visualize ligand-binding sites:
- Example: PyMOL script for site analysis:pythonCopy code
cmd.load("sample.pdb") cmd.select("binding_site", "resi 45-60") cmd.show("surface", "binding_site")
- Example: PyMOL script for site analysis:pythonCopy code
- Deploy workflows on AWS or Google Cloud for flexible scaling.
- Use preconfigured virtual machines with GPU support (e.g., AWS Deep Learning AMIs).
- Submit batch jobs to HPC clusters using SLURM or similar schedulers:
sbatch run_esm3_hpc.sh - Use orchestration tools like Nextflow to manage cloud-based workflows:
- Example Nextflow configuration:nextflowCopy code
process run_esm3 { input: file fasta from "inputs/*.fasta" output: file "outputs/*.pdb" script: """ python examples/run_pretrained_model.py esm2_t6_8M_UR50D --device cuda file done
file done
- Example Nextflow configuration:nextflowCopy code
- Step 3: Identify Functional Domains
- Use tools like HMMER to detect conserved motifs in predicted structures:
hmmsearch --domtblout domains.out pfam.hmm batch_results.fasta
- Use tools like HMMER to detect conserved motifs in predicted structures:
- Step 4: Generate Comprehensive Reports
- Combine predictions and annotations into a unified report:pythonCopy code
import pandas as pd structures = pd.read_csv("predictions.csv") domains = pd.read_csv("domains.out") report = pd.merge(structures, domains, on="sequence_id") report.to_csv("annotation_report.csv", index=False)
- Combine predictions and annotations into a unified report:pythonCopy code
- Annotating unknown proteins in metagenomes.
- Identifying novel enzymes for biotechnological applications.
- Investigating evolutionary relationships across species.
- Step 1: Generate Training Data
- Use ESM3 to predict structures and annotate datasets:
python examples/run_pretrained_model.py esm2_t6_8M_UR50D --output training_data.csv
- Use ESM3 to predict structures and annotate datasets:
- Step 2: Train AI Models
- Train machine learning models using structural features:pythonCopy code
from sklearn.ensemble import RandomForestClassifier model = RandomForestClassifier() model.fit(X_train, y_train)
- Train machine learning models using structural features:pythonCopy code
- Step 3: Predict New Outcomes
- Use the trained model to predict properties of novel sequences:pythonCopy code
predictions = model.predict(X_test)
- Use the trained model to predict properties of novel sequences:pythonCopy code
- Step 4: Validate Predictions
- Compare AI model predictions with experimental data or further ESM3 predictions.
- Predicting protein-drug interactions.
- Designing de novo proteins with desired properties.
- Classifying proteins based on structural or functional similarity.
- As the size of datasets grows, scalability becomes a critical concern. Future updates to ESM3 could incorporate native support for distributed computing.
- Proposed Feature:
- Enable seamless multi-node execution for HPC clusters.
- Impact:
- Accelerates predictions for entire proteomes or metagenomic datasets, making high-throughput studies more feasible.
- Preconfigured instances of ESM3 on platforms like AWS or Google Cloud would simplify accessibility for users lacking local computational resources.
- Impact:
- Democratizes access to ESM3, reducing barriers for resource-constrained researchers.
- Future versions of ESM3 could support multimodal inputs, including RNA sequences, small molecules, and chemical structures.
- Proposed Feature:
- Train models to predict interactions between different biomolecules, such as RNA-protein or protein-ligand complexes.
- Impact:
- Broadens ESM3’s application in systems biology and drug discovery.
- Integration with models capable of generating structural visualizations or molecular descriptions in natural language.
- Impact:
- Enhances interpretability and usability for non-expert users.
- Leveraging ESM3 for precision medicine by predicting the effects of patient-specific mutations on protein function.
- Future Enhancements:
- Incorporate patient-specific datasets to personalize predictions.
- Impact:
- Enables targeted therapy design for genetic diseases or cancer.
- Extend ESM3’s capabilities to identify novel biomarkers by analyzing protein conformational changes under disease conditions.
- Impact:
- Supports early diagnostics and individualized treatment plans.
- ESM3 could be extended to support iterative design workflows that optimize proteins for industrial, therapeutic, or environmental applications.
- Proposed Features:
- Incorporate generative design capabilities to suggest novel protein sequences.
- Impact:
- Revolutionizes the design of synthetic enzymes, biosensors, and drug candidates.
- Improve ESM3’s accuracy in predicting protein stability under extreme conditions, such as high temperatures or acidic environments.
- Impact:
- Expands applications in biotechnology and industrial manufacturing.
- Extend ESM3’s predictions to multi-protein complexes, improving its utility in systems biology.
- Proposed Features:
- Enable simultaneous modeling of multiple interacting proteins.
- Impact:
- Advances research into signaling pathways, protein assembly, and supramolecular structures.
- Develop tools for real-time prediction of protein structures during experimental procedures such as crystallography or cryo-EM.
- Impact:
- Accelerates the pace of structural biology research.
- ESM3 could be adapted to study environmental microbiomes and their role in carbon sequestration, pollution breakdown, or bioenergy production.
- Impact:
- Promotes sustainable solutions for environmental challenges.
- Leverage ESM3’s modeling capabilities to design protein-based materials with unique mechanical, optical, or thermal properties.
- Impact:
- Drives innovation in nanotechnology and advanced materials.
- Develop graphical user interfaces (GUIs) or web-based platforms for ESM3.
- Impact:
- Broadens ESM3’s appeal to non-programmers and interdisciplinary researchers.
- Provide pre-annotated datasets and interactive tutorials to lower the learning curve for new users.
- Impact:
- Encourages widespread adoption among academic and industrial communities.
- Foster a vibrant developer community to contribute new features, models, and tools.
- Proposed Initiative:
- Create a plugin architecture that allows external modules to extend ESM3’s functionality.
- Impact:
- Accelerates innovation and diversification of ESM3 applications.
- Establish standardized datasets and benchmarks for evaluating ESM3’s performance in different applications.
- Impact:
- Ensures transparency and comparability across studies.
- Investigate the integration of quantum computing for solving complex protein folding problems beyond the scope of classical computation.
- Impact:
- Breakthroughs in computational efficiency and accuracy.
- Enable collaborative training of ESM3 models across institutions without sharing sensitive data.
- Impact:
- Enhances model robustness while preserving data privacy.
- ESM3’s ability to predict protein structures and interactions has redefined the boundaries of molecular biology. By providing accurate, high-resolution models:
- Researchers can explore the intricate details of protein folding and dynamics.
- Insights into protein-protein interactions lead to novel therapeutic strategies and drug design approaches.
- Example:
- The application of ESM3 in identifying functional sites within enzymes has enabled the design of bio-catalysts for industrial use.
- ESM3 serves as a versatile tool for addressing challenges in genomics, proteomics, environmental sciences, and materials science. By offering:
- High scalability for large datasets.
- Integration with downstream tools for complex workflows.
- It has become a pivotal asset in interdisciplinary studies that require bridging biology, chemistry, and computational sciences.
- One of ESM3’s defining strengths is its accessibility:
- Open-source nature ensures widespread adoption without financial barriers.
- Pre-trained models allow immediate application to real-world problems without requiring extensive customization.
- Whether analyzing single proteins or entire proteomes, ESM3’s scalable architecture supports a wide range of applications:
- Researchers can tailor workflows to their computational resources, from personal devices to HPC clusters and cloud platforms.
- The incorporation of state-of-the-art transformer-based architectures gives ESM3 its predictive power:
- Achieving a balance between computational efficiency and biological accuracy.
- Providing confidence scores and structural annotations for rigorous scientific interpretation.
- Multi-protein complexes, membrane proteins, and intrinsically disordered regions present modeling difficulties:
- Advances in training datasets and algorithmic improvements will be necessary to address these challenges effectively.
- As ESM3 expands its applications into fields like environmental science and material design:
- Harmonizing workflows with non-biological data and tools will require further refinement.
- Simplified interfaces and user-friendly resources are needed to empower non-experts to fully utilize ESM3’s capabilities.
- Fostering a collaborative community around ESM3 is critical to its evolution:
- Contributions of plugins, workflows, and benchmarks can enhance its versatility.
- Open forums for knowledge exchange will drive innovation.
- By integrating quantum computing, federated learning, and advanced visualization techniques, ESM3 can remain at the forefront of computational tools:
- Expanding its reach into new areas of science and technology.
- From aiding in drug discovery to tackling climate change, ESM3’s impact will grow as researchers find new ways to apply its capabilities:
- Large-scale adoption in clinical settings for personalized medicine.
- Widespread use in industry for sustainable solutions.
- Adopt ESM3 in their workflows to unlock new insights and efficiencies.
- Contribute to its growth through open-source collaboration and shared use cases.
- Educate others on its potential, fostering a global community of users who can leverage ESM3 for societal and scientific advancement.
8.3 Execution Errors
Problem: Invalid Input File Format
Problem: Memory Overflow
8.4 Performance Bottlenecks
Problem: Slow Execution
Problem: High Disk Usage
8.5 Common Output Errors
Problem: Missing or Corrupted Output Files
Problem: Unexpected Results
8.6 Resources for Additional Help
1. Official Documentation
2. Community Support
3. Diagnostic Tools
Troubleshooting is a vital skill for maximizing the utility of ESM3. By systematically diagnosing issues, leveraging detailed error messages, and applying the solutions outlined in this chapter, users can resolve most problems encountered during installation, configuration, and execution. With a fully operational setup, the next chapter will focus on best practices for long-term use and maintenance of ESM3, ensuring consistent performance and adaptability for evolving research needs.
9. Best Practices for Long-Term Use
Proper maintenance and optimization of ESM3 (Evolutionary Scale Modeling 3) are critical for ensuring consistent performance and adapting the tool to evolving research demands. This chapter provides best practices for long-term use, including strategies for managing updates, optimizing workflows, and maintaining a reliable environment for ongoing research.
9.1 Regularly Updating ESM3
ESM3 is actively maintained by its developers, with frequent updates that enhance functionality, improve performance, and address bugs. Staying up-to-date ensures access to the latest features and models.
1. Monitor the Repository for Updates
2. Update the Cloned Repository
3. Reinstall Dependencies After Updates
4. Test After Updates
9.2 Optimizing Workflows for Efficiency
As your research evolves, optimizing ESM3 workflows can save time and computational resources, particularly for large-scale projects.
1. Automate Common Tasks
2. Parallel Execution
10.4 Integration with Molecular Dynamics
ESM3 outputs are highly compatible with molecular dynamics (MD) simulations, enabling refinement of predicted structures.
1. Preparing ESM3 Outputs for MD
2. Running MD Simulations
3. Analyzing MD Results
10.5 Integration with Functional Analysis Tools
Functional analysis of predicted structures reveals insights into protein activity, interaction, and potential applications.
1. Functional Annotation
2. Docking Simulations
3. Binding Site Prediction
10.6 Scaling Workflows with Cloud and HPC
For resource-intensive workflows, cloud computing and high-performance computing (HPC) provide scalability.
1. Cloud Platforms
2. HPC Clusters
3. Workflow Orchestration
2. Applications
11.5 Integrating ESM3 with AI Models
Combining ESM3 with machine learning and deep learning models unlocks powerful predictive capabilities for diverse applications.
1. Workflow for AI Integration
2. Applications
ESM3’s advanced use cases demonstrate its versatility and power in addressing some of the most challenging problems in computational biology and bioinformatics. From modeling protein-protein interactions to designing novel therapeutics, ESM3 provides researchers with the tools needed to achieve significant breakthroughs. By integrating ESM3 into complex workflows and leveraging its outputs for downstream applications, researchers can harness its full potential to push the boundaries of science. The next chapter will focus on future directions, exploring emerging trends and opportunities for further development and application of ESM3.
12. Future Directions for ESM3
As a cutting-edge tool in computational biology and bioinformatics, ESM3 (Evolutionary Scale Modeling 3) continues to evolve, opening new avenues for research and applications across diverse scientific domains. This chapter explores emerging trends, potential advancements, and opportunities for extending the capabilities of ESM3. By identifying future directions, researchers can align their work with the trajectory of ESM3’s development and contribute to its growing impact.
12.1 Enhancing Scalability for Large-Scale Projects
1. Optimizing Performance on High-Performance Computing (HPC) Systems
2. Cloud-Based Implementations
12.2 Integration with Multimodal AI Models
1. Expanding Beyond Protein Sequences
2. Coupling with Vision-Language Models
12.3 Applications in Personalized Medicine
1. Mutational Impact Predictions
2. Predictive Modeling for Biomarker Discovery
12.4 Advancements in Protein Engineering
1. De Novo Protein Design
2. Enhanced Stability Predictions
12.5 Expansion into Structural Biology
1. Modeling Protein Complexes
2. Real-Time Modeling
12.6 Cross-Disciplinary Applications
1. Environmental Sciences
2. Materials Science
12.7 Increasing Accessibility and User Experience
1. Simplified Interfaces
2. Comprehensive Tutorials and Datasets
12.8 Strengthening Community Contributions
1. Open-Source Collaboration
2. Shared Repositories for Benchmarking
12.9 Leveraging Emerging Technologies
1. Quantum Computing
2. Federated Learning
The future of ESM3 is filled with promise, driven by its ability to address complex challenges in computational biology and beyond. By focusing on scalability, interdisciplinary integration, and enhanced usability, ESM3 is poised to become a cornerstone of modern research. Researchers and developers alike are encouraged to contribute to its growth, ensuring that ESM3 remains at the forefront of scientific discovery.
13. Conclusion
The journey through ESM3 (Evolutionary Scale Modeling 3) demonstrates its transformative potential across scientific domains, from protein modeling and structural biology to environmental modeling and personalized medicine. As a tool at the cutting edge of AI and computational biology, ESM3 bridges the gap between raw sequence data and actionable insights, enabling researchers to address complex biological questions with unprecedented precision and scalability.
13.1 The Impact of ESM3
1. Revolutionizing Protein Science
2. Advancing Interdisciplinary Research
13.2 Key Takeaways
1. Accessibility and Usability
2. Scalability
3. Accuracy and Precision
13.3 Remaining Challenges
While ESM3 has proven transformative, challenges remain that require continued innovation and development.
1. Handling Complex Systems
2. Integration Across Disciplines
3. Democratizing Advanced Use
13.4 The Path Forward
1. Community Engagement
2. Emerging Technologies
3. Expanding Real-World Applications
13.5 Call to Action
Researchers, developers, and educators are encouraged to:
ESM3 is more than just a tool; it represents a paradigm shift in computational biology and related disciplines. By merging the power of AI with the intricacies of biological data, ESM3 empowers researchers to tackle some of the most pressing scientific challenges of our time. With ongoing innovation and community support, the possibilities for ESM3’s impact are boundless, setting the stage for a new era of discovery and understanding.
Sample Configuration File
Below is a sample configuration file for running an ESM3 model. Save this as esm3_config.yaml in your project directory.
yaml# ESM3 Configuration File
general:
model_name: "esm2_t6_8M_UR50D" # Pre-trained model to use
device: "cuda" # Specify 'cuda' for GPU or 'cpu' for CPU
input:
input_file: "data/sample.fasta" # Path to input FASTA file
batch_size: 32 # Number of sequences processed in each batch
output:
output_dir: "outputs/" # Directory for saving predictions
log_file: "logs/esm3_run.log" # Log file to capture execution details
advanced:
precision: "fp32" # Floating point precision ('fp16' for faster GPU runs)
max_tokens: 1024 # Maximum tokens per sequence
enable_debug: false # Set to true for verbose debugging information
B. Command Reference Guide
This section provides a list of commonly used commands for running and managing ESM3 models.
1. Running a Pre-Trained Model
Use the following command to run a pre-trained ESM3 model:
python examples/run_pretrained_model.py esm2_t6_8M_UR50D --device cuda data/sample.fasta
2. Specify Output Directory
Customize the directory for storing prediction outputs:
python examples/run_pretrained_model.py esm2_t6_8M_UR50D --device cuda --output_dir outputs/ data/sample.fasta
3. Process Multiple Sequences in Batches
Define batch size for processing larger datasets:
python examples/run_pretrained_model.py esm2_t6_8M_UR50D --device cuda --batch_size 64 data/large_dataset.fasta
4. Enable Debugging
Enable debugging mode to log detailed execution steps:
python examples/run_pretrained_model.py esm2_t6_8M_UR50D --device cuda --debug data/sample.fasta
5. Run on CPU
If GPU is unavailable, specify CPU for execution:
python examples/run_pretrained_model.py esm2_t6_8M_UR50D --device cpu data/sample.fasta
1. Introduction
ESM3 (Evolutionary Scale Modeling 3) has become a transformative force in computational biology, setting new benchmarks in protein structure prediction and functional annotation. By leveraging the latest advancements in transformer-based machine learning models, ESM3 enables researchers to decode the structural and functional mysteries of proteins at a scale and precision previously unattainable. However, unlocking the full potential of ESM3 requires careful installation and configuration, tailored to the specific needs of diverse research applications. This article provides a comprehensive, step-by-step guide to ensure a seamless setup, laying the foundation for efficient and accurate protein modeling.
1.1 The Significance of Proper Installation and Configuration
Installing ESM3 is not just a technical prerequisite—it is a critical process that sets the stage for successful protein modeling and analysis. A properly installed and configured ESM3 ensures:
Proper installation ensures that users can focus on scientific discovery rather than troubleshooting technical issues, maximizing the impact of their research.
1.2 Why Choose ESM3?
Before diving into installation specifics, it’s important to understand the unparalleled advantages that ESM3 offers:
ESM3’s transformative features make it an essential tool for tackling some of the most complex challenges in modern science.
1.3 Challenges in Installing and Configuring ESM3
While ESM3 is designed to be user-friendly and accessible, its installation process can present unique challenges, especially for users without extensive computational experience. Some common obstacles include:
This article is designed to address these challenges comprehensively, offering solutions and best practices tailored to diverse research needs.
1.4 Objectives of This Guide
This tutorial is structured to ensure users achieve a seamless setup of ESM3. By the end of the article, readers will:
This guide ensures a robust and flexible setup, empowering users to leverage ESM3 to its fullest extent.
1.5 Target Audience
This guide is designed for:
Whether you are a seasoned researcher or a first-time user, this guide provides the clarity and depth needed for a successful installation and configuration experience.
1.6 Structure of the Article
To guide users through the installation process, this article is divided into well-defined sections, each addressing a critical aspect of the setup:
1.7 Setting the Foundation for Success
A well-prepared installation and configuration process ensures that users can fully exploit ESM3’s capabilities without technical disruptions. By following this comprehensive guide, users will be equipped with the tools and knowledge to integrate ESM3 seamlessly into their research workflows. From fundamental setup steps to advanced customization, this guide is your roadmap to unlocking the transformative power of ESM3 in protein modeling and beyond.
2. Pre-Installation Checklist
Proper preparation is critical for a successful installation of ESM3 (Evolutionary Scale Modeling 3). Before diving into the installation steps, it is essential to ensure your computational environment meets the necessary requirements and is set up to handle ESM3’s dependencies and workflows. This chapter provides a comprehensive checklist to help users avoid common pitfalls, ensuring a smooth installation process.
2.1 Understanding System Requirements
ESM3 is a resource-intensive tool that performs advanced computational tasks, including large-scale protein modeling and functional annotation. Below are the minimum and recommended hardware and software requirements for running ESM3 efficiently:
Hardware Requirements
Software Requirements
2.2 Preparing the System
To avoid disruptions during installation, ensure your system is prepared with the following steps:
1. Update the Operating System
2. Install Python and Pip
3. Install Git
4. Set Up a Virtual Environment
2.3 Installing Dependencies
To ensure ESM3 functions correctly, install the following dependencies:
1. Core Libraries
2. CUDA Toolkit (For GPU Acceleration)
2.4 Preparing a Workspace
Create a dedicated directory for ESM3 to organize files and outputs efficiently:
2.5 Verifying System Readiness
Before proceeding, verify that your system is ready for ESM3 installation:
2.6 Preparing for Advanced Configurations
If you plan to use advanced configurations, such as cloud deployment or integration with other tools, consider these additional preparations:
A thorough pre-installation preparation is critical to a smooth and successful setup of ESM3. By ensuring your system meets the hardware and software requirements, installing necessary dependencies, and preparing a clean workspace, you reduce the likelihood of errors and optimize ESM3’s performance from the start. With this checklist complete, you are ready to move on to downloading and installing ESM3 with confidence.
3. Downloading ESM3
The first step in the installation process is obtaining the ESM3 software package. As an open-source tool, ESM3 (Evolutionary Scale Modeling 3) is freely available through GitHub. However, downloading and preparing the source code requires attention to detail to ensure that all components are correctly set up. This chapter guides users through the process of accessing the official repository, verifying files, and selecting the best method for their specific needs.
3.1 Accessing the Official Repository
The primary source for ESM3 is the GitHub repository maintained by its developers. The repository includes the source code, installation instructions, and updates. Follow these steps to access it:
3.2 Cloning the Repository
Cloning the repository ensures you have the latest version of ESM3 and simplifies future updates.
3.3 Downloading a Pre-Packaged Release
For users who prefer not to use Git, pre-packaged releases are available on the GitHub repository.
3.4 Verifying File Integrity
To ensure a successful installation, it’s essential to verify that the downloaded files are complete and uncorrupted.
3.5 Choosing the Installation Method
Depending on your computational needs and resources, you can install ESM3 in one of the following ways:
3.6 Preparing for the Next Steps
Before proceeding to installation, ensure you have:
Downloading ESM3 is a straightforward yet crucial step in the installation process. Whether you choose to clone the repository for ongoing updates or download a pre-packaged release for immediate use, following these detailed instructions ensures a reliable starting point. With the files prepared and verified, you’re now ready to proceed to the next phase: installing ESM3 on your system. The upcoming chapter provides a comprehensive guide for setting up ESM3 on Linux, macOS, and Windows systems, tailored to diverse computational environments.
4. Installing ESM3 Locally
Installing ESM3 (Evolutionary Scale Modeling 3) on your local machine involves several steps, tailored to the specific requirements of your operating system. This chapter provides detailed, step-by-step instructions for installing ESM3 on Linux, macOS, and Windows (via Windows Subsystem for Linux, WSL). By carefully following these instructions, you can ensure a successful installation and prepare your system for optimal performance.
4.1 General Preparations for Installation
Before proceeding with installation, confirm the following:
4.2 Installing on Linux
Linux provides a robust platform for running computational tools like ESM3. The installation process is straightforward, provided all dependencies are correctly configured.
Step 1: Navigate to the Repository
Step 2: Install Python Dependencies
Step 3: Install CUDA for GPU Support (Optional)
Step 4: Install ESM3
Step 5: Verify the Installation
4.3 Installing on macOS
macOS users can install ESM3 with Homebrew and Python. GPU acceleration is not natively supported on macOS, but CPU-based installations are fully functional.
Step 1: Navigate to the Repository
Step 2: Install Dependencies
Step 3: Install ESM3
Step 4: Verify the Installation
4.4 Installing on Windows (via WSL)
Windows users can leverage the Windows Subsystem for Linux (WSL) to run a Linux environment, enabling ESM3 installation and use.
Step 1: Set Up WSL
Step 2: Install Dependencies
Step 3: Install CUDA for GPU Support (Optional)
Step 4: Navigate to the Repository
Step 5: Install ESM3
Step 6: Verify the Installation
4.5 Verifying Installation Across Platforms
After completing the installation process, verify that ESM3 is correctly installed and ready for use:
4.6 Post-Installation Steps
After successfully installing ESM3, consider the following actions to optimize your setup:
8.3 Execution Errors
Problem: Invalid Input File Format
Problem: Memory Overflow
8.4 Performance Bottlenecks
Problem: Slow Execution
Problem: High Disk Usage
8.5 Common Output Errors
Problem: Missing or Corrupted Output Files
Problem: Unexpected Results
8.6 Resources for Additional Help
1. Official Documentation
2. Community Support
3. Diagnostic Tools
Troubleshooting is a vital skill for maximizing the utility of ESM3. By systematically diagnosing issues, leveraging detailed error messages, and applying the solutions outlined in this chapter, users can resolve most problems encountered during installation, configuration, and execution. With a fully operational setup, the next chapter will focus on best practices for long-term use and maintenance of ESM3, ensuring consistent performance and adaptability for evolving research needs.
9. Best Practices for Long-Term Use
Proper maintenance and optimization of ESM3 (Evolutionary Scale Modeling 3) are critical for ensuring consistent performance and adapting the tool to evolving research demands. This chapter provides best practices for long-term use, including strategies for managing updates, optimizing workflows, and maintaining a reliable environment for ongoing research.
9.1 Regularly Updating ESM3
ESM3 is actively maintained by its developers, with frequent updates that enhance functionality, improve performance, and address bugs. Staying up-to-date ensures access to the latest features and models.
1. Monitor the Repository for Updates
2. Update the Cloned Repository
3. Reinstall Dependencies After Updates
4. Test After Updates
9.2 Optimizing Workflows for Efficiency
As your research evolves, optimizing ESM3 workflows can save time and computational resources, particularly for large-scale projects.
1. Automate Common Tasks
2. Parallel Execution
10.4 Integration with Molecular Dynamics
ESM3 outputs are highly compatible with molecular dynamics (MD) simulations, enabling refinement of predicted structures.
1. Preparing ESM3 Outputs for MD
2. Running MD Simulations
3. Analyzing MD Results
10.5 Integration with Functional Analysis Tools
Functional analysis of predicted structures reveals insights into protein activity, interaction, and potential applications.
1. Functional Annotation
2. Docking Simulations
3. Binding Site Prediction
10.6 Scaling Workflows with Cloud and HPC
For resource-intensive workflows, cloud computing and high-performance computing (HPC) provide scalability.
1. Cloud Platforms
2. HPC Clusters
3. Workflow Orchestration
2. Applications
11.5 Integrating ESM3 with AI Models
Combining ESM3 with machine learning and deep learning models unlocks powerful predictive capabilities for diverse applications.
1. Workflow for AI Integration
2. Applications
ESM3’s advanced use cases demonstrate its versatility and power in addressing some of the most challenging problems in computational biology and bioinformatics. From modeling protein-protein interactions to designing novel therapeutics, ESM3 provides researchers with the tools needed to achieve significant breakthroughs. By integrating ESM3 into complex workflows and leveraging its outputs for downstream applications, researchers can harness its full potential to push the boundaries of science. The next chapter will focus on future directions, exploring emerging trends and opportunities for further development and application of ESM3.
12. Future Directions for ESM3
As a cutting-edge tool in computational biology and bioinformatics, ESM3 (Evolutionary Scale Modeling 3) continues to evolve, opening new avenues for research and applications across diverse scientific domains. This chapter explores emerging trends, potential advancements, and opportunities for extending the capabilities of ESM3. By identifying future directions, researchers can align their work with the trajectory of ESM3’s development and contribute to its growing impact.
12.1 Enhancing Scalability for Large-Scale Projects
1. Optimizing Performance on High-Performance Computing (HPC) Systems
2. Cloud-Based Implementations
12.2 Integration with Multimodal AI Models
1. Expanding Beyond Protein Sequences
2. Coupling with Vision-Language Models
12.3 Applications in Personalized Medicine
1. Mutational Impact Predictions
2. Predictive Modeling for Biomarker Discovery
12.4 Advancements in Protein Engineering
1. De Novo Protein Design
2. Enhanced Stability Predictions
12.5 Expansion into Structural Biology
1. Modeling Protein Complexes
2. Real-Time Modeling
12.6 Cross-Disciplinary Applications
1. Environmental Sciences
2. Materials Science
12.7 Increasing Accessibility and User Experience
1. Simplified Interfaces
2. Comprehensive Tutorials and Datasets
12.8 Strengthening Community Contributions
1. Open-Source Collaboration
2. Shared Repositories for Benchmarking
12.9 Leveraging Emerging Technologies
1. Quantum Computing
2. Federated Learning
The future of ESM3 is filled with promise, driven by its ability to address complex challenges in computational biology and beyond. By focusing on scalability, interdisciplinary integration, and enhanced usability, ESM3 is poised to become a cornerstone of modern research. Researchers and developers alike are encouraged to contribute to its growth, ensuring that ESM3 remains at the forefront of scientific discovery.
13. Conclusion
The journey through ESM3 (Evolutionary Scale Modeling 3) demonstrates its transformative potential across scientific domains, from protein modeling and structural biology to environmental modeling and personalized medicine. As a tool at the cutting edge of AI and computational biology, ESM3 bridges the gap between raw sequence data and actionable insights, enabling researchers to address complex biological questions with unprecedented precision and scalability.
13.1 The Impact of ESM3
1. Revolutionizing Protein Science
2. Advancing Interdisciplinary Research
13.2 Key Takeaways
1. Accessibility and Usability
2. Scalability
3. Accuracy and Precision
13.3 Remaining Challenges
While ESM3 has proven transformative, challenges remain that require continued innovation and development.
1. Handling Complex Systems
2. Integration Across Disciplines
3. Democratizing Advanced Use
13.4 The Path Forward
1. Community Engagement
2. Emerging Technologies
3. Expanding Real-World Applications
13.5 Call to Action
Researchers, developers, and educators are encouraged to:
ESM3 is more than just a tool; it represents a paradigm shift in computational biology and related disciplines. By merging the power of AI with the intricacies of biological data, ESM3 empowers researchers to tackle some of the most pressing scientific challenges of our time. With ongoing innovation and community support, the possibilities for ESM3’s impact are boundless, setting the stage for a new era of discovery and understanding.
Sample Configuration File
Below is a sample configuration file for running an ESM3 model. Save this as esm3_config.yaml in your project directory.
yaml# ESM3 Configuration File
general:
model_name: "esm2_t6_8M_UR50D" # Pre-trained model to use
device: "cuda" # Specify 'cuda' for GPU or 'cpu' for CPU
input:
input_file: "data/sample.fasta" # Path to input FASTA file
batch_size: 32 # Number of sequences processed in each batch
output:
output_dir: "outputs/" # Directory for saving predictions
log_file: "logs/esm3_run.log" # Log file to capture execution details
advanced:
precision: "fp32" # Floating point precision ('fp16' for faster GPU runs)
max_tokens: 1024 # Maximum tokens per sequence
enable_debug: false # Set to true for verbose debugging information
B. Command Reference Guide
This section provides a list of commonly used commands for running and managing ESM3 models.
1. Running a Pre-Trained Model
Use the following command to run a pre-trained ESM3 model:
python examples/run_pretrained_model.py esm2_t6_8M_UR50D --device cuda data/sample.fasta
2. Specify Output Directory
Customize the directory for storing prediction outputs:
python examples/run_pretrained_model.py esm2_t6_8M_UR50D --device cuda --output_dir outputs/ data/sample.fasta
3. Process Multiple Sequences in Batches
Define batch size for processing larger datasets:
python examples/run_pretrained_model.py esm2_t6_8M_UR50D --device cuda --batch_size 64 data/large_dataset.fasta
4. Enable Debugging
Enable debugging mode to log detailed execution steps:
python examples/run_pretrained_model.py esm2_t6_8M_UR50D --device cuda --debug data/sample.fasta
5. Run on CPU
If GPU is unavailable, specify CPU for execution:
python examples/run_pretrained_model.py esm2_t6_8M_UR50D --device cpu data/sample.fasta
Leave a Reply